Last Updated: January 28, 2026
Introduction to Vitamin B3: Nicotinic Acid/Niacin
Vitamin B3 is nicotinic acid, more commonly called niacin. Both nicotinic acid (NA) and nicotinamide (NAM) can serve as the dietary source of the co-factor forms of vitamin B3, nicotinamide adenine dinucleotide (NAD+) and nicotinamide adenine dinucleotide phosphate (NADP+).

Synthesis and Salvage of NAD+
With the exception of neurons, human cells cannot transport NAD+ into the cell, therefore, they must synthesize it de novo or salvage it. The pathway of de novo synthesis involves the amino acid tryptophan. However, due to the critical need for NAD+, most cells recycle the cofactor via salvage pathways as opposed to de novo synthesis. The salvage pathways for NAD+ synthesis take place in both the nucleus and the mitochondria and involve either nicotinic acid (NA), nicotinamide (NAM), or nicotinamide riboside (NR).
Synthesis of NAD+ from nicotinamide (NAM) or from nicotinamide riboside (NR) occurs in a two-step processes. Synthesis of NAD+ from nicotinic acid (NA) occurs in a three-step process. The three-step pathway of NAD+ synthesis from NA is referred to as the Preiss-Handler pathway.
The de novo pathway of NAD+ synthesis from tryptophan and the salvage pathways from NAM and from NA each requires the activated form of ribose, 5-phosphoribosyl 1-pyrophosphate (PRPP). PRPP is the same activated ribose required for nucleotide synthesis.
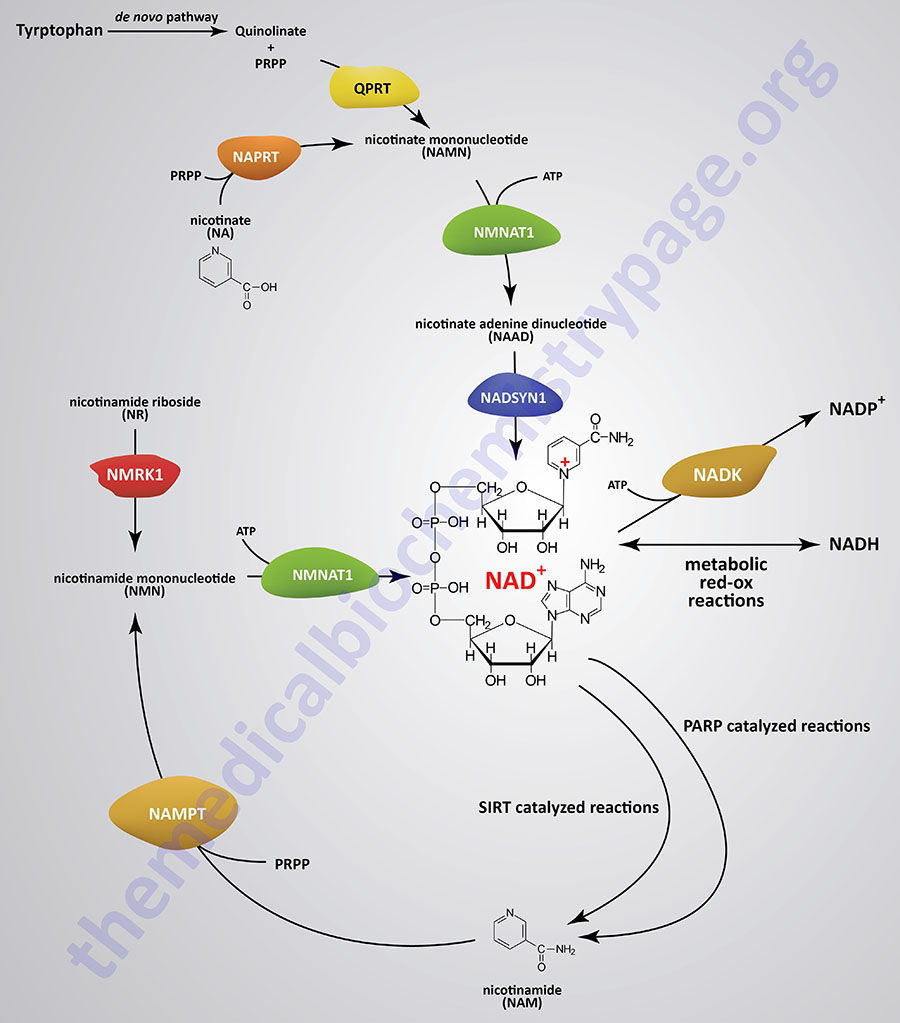
NAD+ Synthesis from Tryptophan
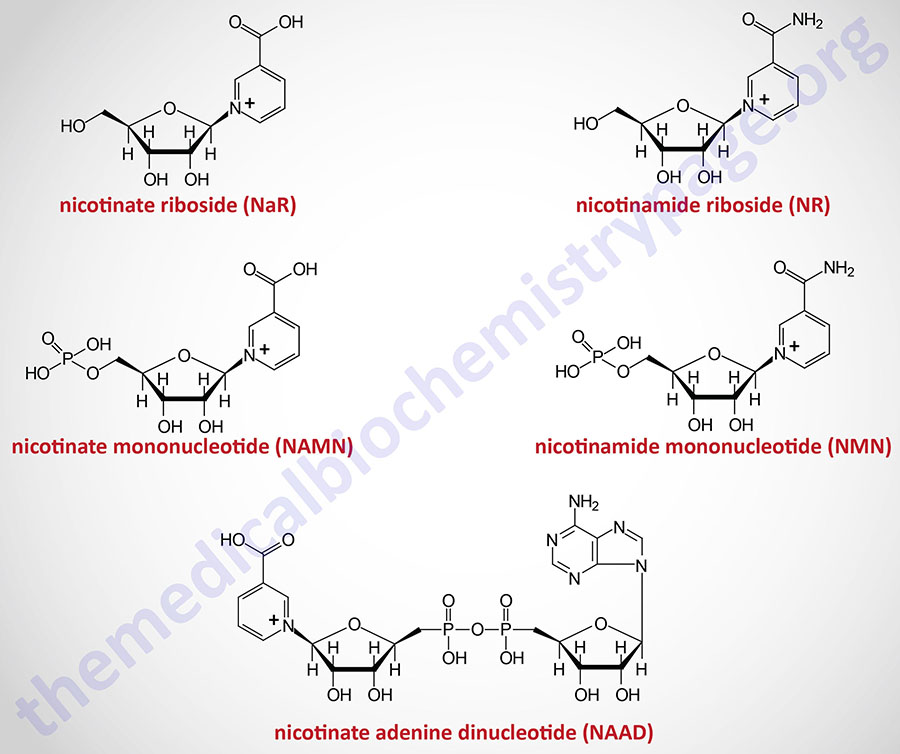
The details of the pathway of de novo NAD+ synthesis from the amino acid tryptophan are covered in the Amino Acid Catabolism page. The conversion of tryptophan to NAD+ occurs predominantly in the liver and utilizes the metabolite of tryptophan catabolism, kynurenine. Synthesis of NAD+ has also been shown to occur within the kidneys, not only from tryptophan-derived kynurenine, but also from hepatocyte-derived nicotinic acid riboside (NaR; also designated NAR).
NAD+ Salvage from Nicotinic Acid (NA)
Nicotinic acid (NA) is converted to nicotinate mononucleotide (NAMN) via the action of nicotinate phosphoribosyltransferase which is encoded by the NAPRT gene.
The NAPRT gene is located on chromosome 8q24.3 and is composed of 13 exons that generate four alternatively spliced mRNAs each encoding distinct isoforms of the enzyme.
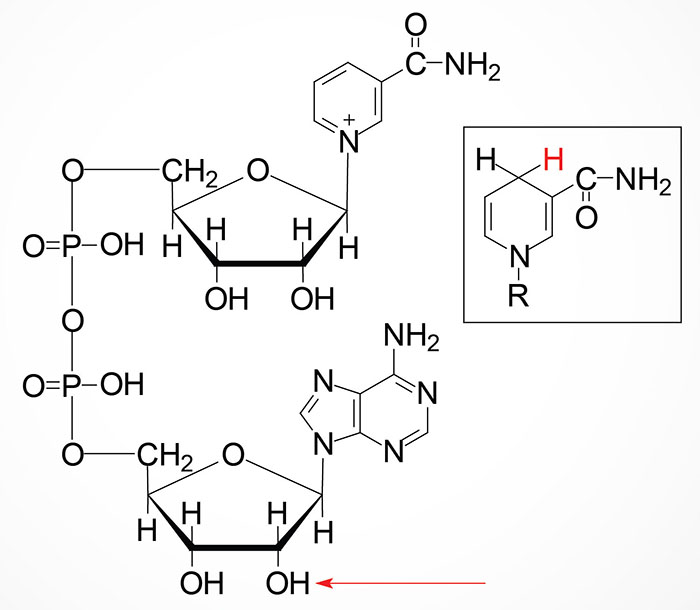
NAMN is then converted to nicotinate adenine dinucleotide (NAAD) by the action of the NMNAT enzymes. NAAD is then converted to NAD+ via the action of NAD synthetase 1 (also called glutamine-dependent NAD synthetase) which is encoded by the NADSYN1 gene.
The NADSYN1 gene is located on chromosome 11q13.4 and is composed of 21 exons that encode a 706 amino acid protein.
NAD+ Salvage from Nicotinamide (NAM)
Nicotinamide (NAM) is one form of vitamin B3 found in fortified foods but it is only found at trace levels naturally in foods consumed by humans. Nicotinamide conversion to NAD+ represents one of the salvage pathway to NAD+ synthesis. The primary source of NAM is as a byproduct of the enzymes [e.g. poly-ADP-ribose polymerases (PARP) and sirtuin (SIRT) proteins] that use NAD+ in the course of their reactions. This salvage pathway converts NAM to nicotinamide mononucleotide (NMN) via the action of the enzyme, nicotinamide phosphoribosyltransferase, NAMPT.
Nicotinamide phosphoribosyltransferase is encoded by the NAMPT gene. The NAMPT gene is located on chromosome 7q22.3 and is composed of 13 exons that encode a 491 amino acid precursor protein.
Interestingly there was an activity that was identified at high levels in white adipose tissue that was purported to have insulin mimetic effects. This purported activity was originally called visfatin. The protein identified as visfatin was subsequently discovered to be encoded by the NAMPT gene. The level of NAMPT activity changes under different dietary states such as nutrient status, stress, and during exercise. Of clinical significance is the fact that NAMPT levels are reduced in obesity decreasing its role in stress responses. The NAMPT protein is found both intracellularly (identified as iNAMPT) and extracellularly (identified as eNAMPT). Among numerous functions, the activity of NAMPT is required for the regulation of NAD+-dependent deacetylases of the sirtuin (SIRT) family. SIRT enzymes are critical in the regulation of gene expression through their ability to deacetylate histone proteins, thereby altering chromatin structure.
Following the synthesis of NMN, it is then converted to NAD+ through the action of the nicotinamide nucleotide adenylyltransferase (NMNAT) enzymes and ATP. Humans express three NMNAT genes identified as NMNAT1, NMNAT2, and NMNAT3.
The NMNAT1 gene is located on chromosome 1p36.22 and is composed of 9 exons that generate three alternatively spliced mRNAs that collectively encode two distinct protein isoforms. The NMNAT1 encoded proteins are localized to the nucleus.
The NMNAT2 gene is located on chromosome 1q25.3 and is composed of 12 exons that generate two alternatively spliced mRNAs both of which encode distinct protein isoforms. Expression of the NMNAT2 gene is predominantly localized to the brain. The NMNAT2 encoded proteins are found in the cytosol and in the Golgi.
The NMNAT3 gene is located on chromosome 3q23 and is composed of 10 exons that generate 22 alternatively spliced mRNAs collectively encode eight different protein isoforms. The NMNAT3 encoded proteins are localized to the mitochondria.
The distinct localization of the various NMNAT enzymes indicates that there are likely to be distinct pools of NAD+ in the different subcellular organelles and that these pools are independently regulated through the activity of a given NMNAT.
NAD+ Salvage from Nicotinamide Riboside (NR)
Nicotinamide riboside occurs naturally in cow’s milk and numerous yeast-containing foods. Nicotinamide riboside (NR) is converted to nicotinamide mononucleotide (NMN) via the actions of nicotinamide riboside kinase, NMRK. Humans express two NMRK expressing genes, NMRK1 and NMRK2.
The NMRK1 gene is located on chromosome 9q21.13 and is composed of 12 exons that generate four alternatively spliced mRNAs that collectively encode three distinct protein isoforms. Expression of the NMRK1 gene is ubiquitous.
The NMRK2 gene is located on chromosome 19p13.3 and is composed of 8 exons that generate five alternatively spliced mRNAs, each of which encodes a distinct protein isoform. Expression of the NMRK2 gene is exclusive to the heart and its level of expression increases in response to heart failure such as in dilated cardiomyopathy.
The NMN formed from NR is converted to NAD+ via the same enzymes described in the previous section detailing the salvage of NAM.
NAD+ Salvage from Trigonelline
Trigonelline (1-methyl nicotinic acid) is a naturally occurring N-methylated form of nicotinic acid that is found in numerous plant species, is produced by gut microbiota, and is a product of endogenous metabolism in humans. The mechanism by which trigonelline is demethylated prior to utilization is currently unknown. Trigonelline serves as a source of NAD+ primarily via the Preiss-Handler pathway similar to that of nicotinic acid (NA). However, in mice in which the NAPRT gene has been knocked-out, trigonelline can still contribute to the pool of NR and NMN indicating that other mechanisms allow for trigonelline to contribute to NAD+ synthesis.
1-Methyl Nicotinamide (MNA)
1-Methyl nicotinamide (MNA; also as 1-MNA) is structurally related to trigonelline and as such is also known as trigonellamide. The formation of MNA represents a form of nicotinamide (NAM) catabolism that occurs predominantly in the liver. Nicotinamide is converted to NMA and S-adenosylhomocysteine (SAH) via the actions of the S-adenosylmethionine-dependent enzyme, nicotinamide N-methyltransferase (NNMT). In addition to NAM, the action of NNMT include the metabolism of drugs and other xenobiotic compounds in the liver. Following conversion of NAM to MNA the compound is excreted in the urine.
As a result of the activity of NNMT, not just due to its role in methylating NAM, the enzyme plays an important role in the regulation of the overall levels of S-adenosylmethionine (SAM of AdoMet) within cells in addition to the regulation of the SAM to SAH ratio. The actions of NNMT not only consume SAM, but via the interactions of NNMT with methionine synthase and betaine-homocysteine S-methyltransferase (BHMT), the enzyme promotes the regeneration of SAM from homocysteine within the context of the methionine cycle. The activities of NNMT, in the context of NAM metabolism, indicate that the enzyme has a crucial role the regulation of NAD+-dependent signaling as well as methionine metabolism via the consequences of the and simultaneous depletion of NAM and SAM.
Nicotinamide N-methyltransferase is encoded by the NNMT gene. The NNMT gene is located on chromosome 11q23.2 and contains 8 exons that generate four alternatively spliced mRNAs, each of which encode the same 264 amino acid protein. Although expression of the NNMT gene is highest in the liver, expression is found in numerous other tissues such as the heart and adipose tissue.
Although there is evidence to suggest that MNA serves an antioxidant and anti-inflammatory function, there is a negative correlation between the serum levels of MNA in obese individuals and in type 2 diabetes. In addition, when the level of NNMT expression is reduced, such as in gene knock-out mice, there is a consequent protection against diet-induced obesity. In these types of experiments there is also increased insulin sensitivity and a consequent reduction in plasma glucose levels.
Renal Synthesis of NAD+
Hepatocytes can convert nicotinate mononucleotide (NAMN, or NaMN), generated in the context of the Preiss-Handler pathway, to nicotinic acid riboside (NaR, or NAR). This conversion is catalyzed by the cytosolic 5′-nucleotidase II enzyme which is encoded by the NT5C2 gene. The liver then releases NaR to the blood from which it is delivered to the kidneys. The kidneys then convert NaR back to NAMN through the actions of nicotinamide riboside kinase 1 encoded by the NMRK1 gene (also identified as the NRK1 gene). The remainder of the renal NAD+ synthesis pathway is the same as occurs within hepatocytes as diagrammed in the Figure above.
The significance of the renal synthesis of NAD+ is that , like the liver, the kidneys can contribute to circulating levels of nicotinamide that is ultimately utilized for systemic NAD+ synthesis. In addition, experiments in mice have demonstrated that dietary supplementation with NaR results in improvement in whole body NAD+ levels, which normally decline with aging, as well as improving overall kidney health.
Synthesis of NADP+
NAD+ can be converted to NADP+ through the action of cytoplasmic NAD kinase (encoded by the NADK gene) or mitochondrial NAD kinase 2 (encoded by the NADK2 gene).
The NADK gene is located on chromosome 1p36.33 and is composed of 19 exons that generate six alternatively spliced mRNAs that collectively generate four distinct isoforms of the enzyme.
The NADK2 gene is located on chromosome 5p13.2 and is composed of 15 exons that generate four alternatively spliced mRNAs, that collectively encode three distinct isoforms of the enzyme. Expression of the NADK2 gene is highest in the liver, being 5-6 fold higher there than in any other tissue.
NAD+ Degradation
The consumption of NAD+ via the SIRT and PARP family enzymes is considered a form of NAD+ degradation despite the ability of cells to salvage the resultant NAM. NAD+, as well as NADP+, can also be degraded in cells via the actions of glycohydrolases that are members of the ADP-ribosyl cyclase family. These consumption reactions are distinct from the conversion of NAD+ to NADH or to NADP+ or to the release of NAM in the context of the use of NAD+ by SIRT or PARP enzymes.
The NAD+/NADP+ glycohydrolases are encoded by the CD38, BST1 (bone marrow stromal cell antigen 1), and SARM1 (sterile alpha and TIR motif containing 1) genes. TIR refers to Toll/interleukin receptor.
The ADP-ribosyl cyclase activity of these enzymes convert NAD+ and NADP+ to adenosine diphosphoribose (ADPR), cyclic ADP-ribose (cADPR), and nicotinate adenine dinucleotide phosphate (NAADP). The generation of cADPR and NAADP, via the action of these ADP-ribosyl cyclases, represents an important signaling reaction since both molecules can bind to intracellular Ca2+ channels leading to release of stored intracellular Ca2+.
The glycohydrolase activity of these enzymes converts NAD+ to ADPR and nicotinamide (NAM). The NAM can then be recycled to NAD+ via the salvage reactions described above.
The CD38 gene is ubiquitously expressed and the encoded enzyme is the major extracellular NAD+ degrading enzyme in mammals. As humans age the level of CD38 expression increases leading to a concurrent decline in cellular NAD+ levels.
Expression of the BST1 gene is restricted to lymphoid cells and cells of the gut. The BST1 encoded protein is anchored to the membrane via a GPI linkage. The protein encoded by the BST1 gene is also identified as CD157.
Expression of SARM1 is enriched in neural tissues and the protein functions intracellularly. The SARM1 protein can dimerize via the interactions of the TIR motif domains and the dimerized enzyme degrades NAD+, an activity that is associated with destruction of neurons.
Mitochondrial NAD+
Whereas the localization of the NMNAT3 encoded protein is in the mitochondria, little, if any, synthesis or salvage of NAD+ has been shown to take place within the mitochondria itself. Given that ~70% of the total pool of NAD+ is found in the mitochondria, where it functions in the TCA cycle and in electron transport through complex I of the oxidative phosphorylation pathway, its import into this organelle is critical.
Only recently has the mitochondrial NAD+ import transporter been identified. The protein encoded by the SLC25A51 gene is localized to the inner mitochondrial membrane and is required for transport of NAD+ into the matrix of the mitochondria. The SLC25 family of transporters is known as the mitochondrial carrier family. The SLC25A51 encoded protein is also known as mitochondrial carrier triple repeat protein 1, MCART1.
In experiments in cells where the function of SLC25A51 has been ablated, the level of mitochondrial TCA cycle activity is greatly reduced, the level of mitochondrial respiration and electron transport through complex I is greatly reduced, and the levels of mitochondrial NAD+ and NADH are also significantly reduced. These results indicate a critical role for the SLC25A51 encoded transporter in mitochondrial import of NAD+.
Biological Functions of NAD+ and NADP+
NAD+ functions as a cofactor for numerous dehydrogenases including the critical thiamine-dependent enzymes, the pyruvate dehydrogenase complex (PDHc), 2-oxoglutarate (α-ketoglutarate) dehydrogenase (OGDH), and branched-chain α-ketoacid dehydrogenase (BCKD) as well as lactate dehydrogenase and malate dehydrogenase, for example.
NADPH serves as a cofactor for numerous reductive biosynthetic processes including fatty acid synthesis, cholesterol biosynthesis, steroid hormone biosynthesis, and bile acid synthesis.
In total NAD+/NADP+ is required for over 500 enzymatic reactions and as such plays a critical role in the regulation of almost all major biological processes.
Outside the context of metabolic pathways that utilize NAD+ or NADP+ in red-ox reactions, NAD+ is required for a wide array of biological processes. There are two major cell signaling pathways that utilize NAD+ as a co-substrate. The sirtuin proteins and the poly-ADP-ribose polymerases (PARP) are all NAD+-dependent enzymes that function in processes that include mitochondrial metabolism, inflammation, meiosis, autophagy, apoptosis, transcriptional regulation, RNA biogenesis and processing, DNA damage repair, chromatin remodeling, the unfolded protein response (UPR), and circadian rhythms.
The major function of the SIRT proteins is the NAD+-dependent deacetylation of lysine residues in target proteins, particularly histone proteins. During the course of the deacetylation reaction NAM is released and salvaged for synthesis of NAD.
The PARP family of enzymes is encoded by 17 different genes in humans. This family of NAD+-dependent enzymes includes both poly-ADP-ribose polymerases (encoded by the PARP1, 2, and 5 genes) and mono-ADP-ribose transferases (encoded by the PARP3, 4, 6, 10, and 14–16 genes). The reaction catalyzed by PARP enzymes is the cleavage of NAD+ with the subsequent transfer of ADP-ribose to aspartic acid, glutamic acid, and lysine residues on target proteins with the release of NAM.
In addition to the PARP family of ADP-ribosylating enzymes, the SIRT4 and SIRT6 encoded enzymes and the arginine-specific ecto-enzymes (ARTC) also possess ADP-ribosylating activity. The process of ADP-ribosylation is a reversible post-translational modification and it’s presence in a protein is recognized by specific binding proteins termed ADP-ribose “readers”.
Therapeutic Potential for NAD+
Numerous studies have demonstrated an association between the processes of aging and several pathological conditions affecting skeletal muscle, kidney, heart, central nervous system, vision, and hearing with declining levels of NAD+.
The enzyme, quinolinic acid phosphoribosyltransferase (QPRT) is required for tryptophan conversion to NAD+ and reduced levels of this enzyme are associated with acute kidney injury. When patients are given nicotinamide prior to cardiac surgery there is a dose related increase in circulating NAD+ metabolites and a reduction in acute kidney injury in these patients.
High-risk patients with non-melanoma skin cancer have shown reduced incidences of both basal cell carcinomas and squamous cell carcinomas when taking 500 mg of nicotinamide daily for at least 12 months.
Although NAD+ replacement in laboratory animals has shown correlation to reductions in pathology, several studies using nicotinamide riboside (NR) failed to demonstrate functional benefit in obese individuals or the elderly. However, studies using nicotinic acid or nicotinamide have shown the potential for replacement therapies involving NAD+ and its precursors. Given that vitamin B3 can exist as nicotinic acid (niacin), nicotinamide, nicotinamide riboside (NR), and nicotinamide mononucleotide (NMN), all of which are capable of supporting NAD+ synthesis, it is not yet clear whether any one of these NAD+ precursors is superior as an NAD+-replacement therapeutic.
Recent experiments (2025) carried out in laboratory animals have found that dietary supplementation with nicotinic acid (nicotinate) riboside (NaR) leads to improvement in whole body NAD+ levels, particularly during the aging process which is known to be associated with declining levels of NAD+. The kidneys play an important role in this process. In addition, NaR supplementation improves kidney function by supporting the production of NAD+ as well as ATP levels in the kidneys. NaR supplementation also increases NAD+ and ATP levels in the heart in models of aging.