Last Updated: February 19, 2025
Introduction to Mechanistic Target of Rapamycin: mTOR
A central control complex that allows cells to respond and adapt to their particular environment is the Ser/Thr kinase identified as mTOR (mechanistic target of rapamycin). mTOR is the principal component of both the mTOR complex 1 (mTORC1) and mTORC2 protein complexes. mTOR was originally referred to as mammalian target of rapamycin.
The original designation for the protein now identified as mTOR was FRAP1 which stood for FK506-binding protein 12-Rapamycin-Associated Protein 1. Rapamycin was originally isolated and characterized as a naturally occurring anti-fungal compound isolated from Streptomyces hygroscopicus on Easter Island in the south Pacific. Easter Island is called Rapa Nui by the inhabitants of the island, hence the naming of the compound as rapamycin. All drugs isolated from strains of Streptomyces contain the suffix “mycin”.
Following its initial characterization it was discovered that rapamycin exhibited potent immunosuppressive and anti-cancer activity. The immunosuppressive effects of rapamycin are due to it binding immunophilins, the same targets as the immunosuppressive drug FK506 (drug name is tacrolimus; trade name is Prograf). Because the immunophilins, to which rapamycin and tacrolimus interact, bind to FK506 these immunophilins are termed FK506 binding proteins, FKBP. The FKBP were originally defined by their size, such as FKBP-12. FKBP-12 is now identified as FKBP1A. The FKBP are peptidylprolyl cis-trans isomerases. The interaction of rapamycin, or tacrolimus, to FKBP1A enhances the formation of an FKBP1A-FK506 complex which results in increased immunosuppressive effects.
mTOR Complex 1: mTORC1
The mTORC1 is the major mTOR-containing complex that regulates cellular responses to growth factor signaling, nutrient deprivation, and various forms of cellular stress. A major contributor to the activation of the mTORC1 are amino acids which promote the migration of the complex to membranes of the late endosomes and lysosomes (LEL) which represents the major site of mTORC1 activation. Amino acids positively regulate the activity of mTORC1 and during periods of nutrient deprivation, where amino acid levels fall, the level of mTORC1 activity also falls.
In addition to amino acids and growth factor signaling, mTORC1 is activated by oxygen levels and levels of cellular energy. The major kinase that is responsive to energy levels is AMP-activate protein kinase, AMPK. AMPK in turn phosphorylates mTOR within the mTORC1 resulting in activation of the complex.
When the mTORC1 is activated it stimulates numerous anabolic processes while inhibiting catabolic processes. With respect to anabolism, mTORC1 stimulates protein synthesis, nucleotide synthesis, ribosome biogenesis, transcription and processing of mRNAs, lipid biosynthesis, cell growth, adipogenesis, and mitochondrial metabolism. With respect to inhibition of catabolism the mTORC1 inhibits lipid oxidation as well as autophagy.
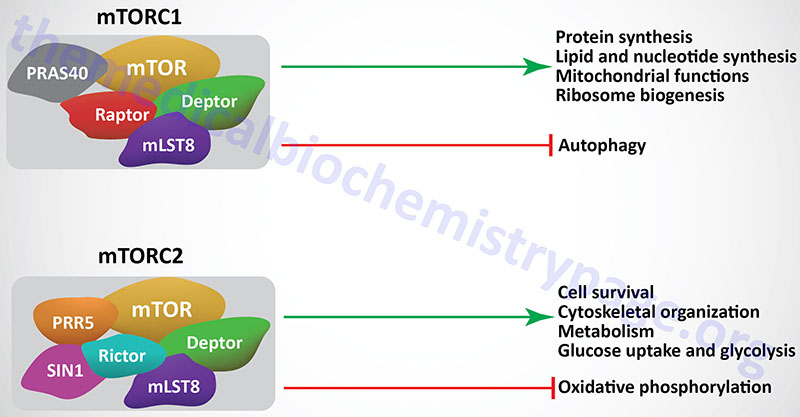
The mTORC1 is composed of mTOR, Raptor, mTOR associated protein yeast LST8 homolog (mLST8), Deptor, and PRAS40.
The mTOR protein is encoded by the MTOR gene. The MTOR gene is large gene located on chromosome 1p36.22 and is composed of 60 exons that generate three alternatively spliced mRNAs encoding proteins of 2549 amino acids (isoform 1) and 2133 amino acids (isoform 2). The gene (ANGPTL7) that encodes the angiopoietin like protein 7 protein is located within one of the introns of the MTOR gene.
The name Raptor is derived from Regulatory Associated Protein of mTOR complex 1 and the protein is encoded by the RPTOR gene. The RPTOR gene is located on chromosome 17q25.3 and is composed of 34 exons that generate two alternatively spliced mRNAs encoding proteins of 1335 amino acids (isoform 1) and 1177 amino acids (isoform 2).
The mLST8 protein is also called mammalian lethal with SEC13 protein 8 and is encoded by the MLST8 gene. The MLST8 encoded protein is also known as GβL (G protein β-subunit-Like protein, pronounced “Gable”). The MLST8 gene is located on chromosome 16p13.3 and is composed of 9 exons that generate seven alternatively spliced mRNAs that collectively encode four distinct protein isoforms.
The name Deptor is derived from DEP domain containing mTOR interacting protein which is encoded by the DEPTOR gene. The DEPTOR gene is located on chromosome 8q24.12 and is composed of 9 exons that generate two alternatively spliced mRNAs encoding proteins of 409 amino acids (isoform 1) and 308 amino acids (isoform 2).
The DEP domain is a globular domain consisting of around 80 amino acids whose name is derived from the three proteins in which it was originally identified (Dishevelled, EGL-10, and Pleckstrin). In humans the EGL-10 homolog is encoded by the RGS7 (regulator of G-protein signaling 7) gene.
The name PRAS40 is derived from Protein-Rich AKT Substrate 40 kDa which was originally isolated from the fruit fly, D. melanogaster. PRAS40 in humans is correctly identified as AKT1 substrate 1 which is encoded by the AKT1S1 gene.
mTOR Complex 2: mTORC2
The mTORC2 is the major mTOR-containing complex that regulates the activity of the kinase AKT/PKB as well as the signaling through the insulin receptor. mTORC2 is also involved in the regulation of the actin containing cytoskeleton. Unlike the inhibition of the activity of mTORC1 by rapamycin, the mTORC2 is not allosterically inhibited by rapamycin.
The mTORC2 is composed of mTOR, DEPTOR, Rictor, SIN1, PRR5, and mLST8. The activities and functions of the mTORC2 are less well characterized in comparison to the mTORC1.
Rictor is Rapamycin-insensitive companion of mTOR which is encoded by the RICTOR gene. The RICTOR gene is located on chromosome 5p13.1 and is composed of 41 exons that generate three alternatively spliced mRNAs, each of which encode a distinct protein isoform.
SIN1 is Stress-activated protein kinase INteracting protein 1. The SIN1 protein is encoded by the MAPKAP1 (MAPK associated protein 1) gene. The MAPKAP1 gene is located on chromosome 9q33.3 and is composed of 13 exons that generate six alternatively spliced mRNAs that collectively encode five distinct protein isoforms.
PRR5 is PRoline Rich 5 which is encoded by the PRR5 gene. The PRR5 encoded protein is also referred to as protein observed with rictor 1 (Proctor1). The PRR5 gene is located on chromosome 22q13.31 and is composed of 11 exons that generate six alternatively spliced mRNAs that collectively encode four distinct protein isoforms.
Function and Regulation of mTORC1 Components
As indicated, the mTOR protein of the mTOR complexes functions as a Ser/Thr kinase. The regulation of the kinase activity of mTOR is controlled by numerous factors including other kinases that phosphorylate mTOR as described in more detail in the following sections. In addition to phosphorylation, the activity of mTOR is regulated by ubiquitylation and by lysine malonylation. Ubiquitylation of mTOR results in its degradation as is typical for the majority of protein ubiquitylation events. The malonylation of mTOR occurs in conjunction with states that result in inhibition of fatty acid synthase (FAS). Malonylation of mTOR results in the inhibition of its kinase activity.
The Raptor protein serves as a critical scaffolding protein allowing for the recruitment of mTOR substrates to the complex. Raptor also undergoes phosphorylation, some of which function in a positive way and some of which function negatively. Numerous kinases have been identified that phosphorylate Raptor including AMPK, the MAP kinase family kinases JNK and p38, NLK (Nemo-like kinase), the large tumor suppressor kinase 1 (LATS1) and LATS2, and by TBK1 (TANK-binding kinase. The activity of Raptor is also regulated by acetylation. When acetylated on Lys1097 (K1097) Raptor enhances the activity of mTORC1.
The DEPTOR protein functions as an inhibitor of the kinase activity of mTOR, thus it inhibits both mTORC1 and mTORC2. The stability of Deptor and its ability to interact with and inhibit mTOR is controlled by its state of phosphorylation. Phosphorylation of Deptor has been shown to occur on at least 14 different Ser residues and two Thr residues. Deptor activity is also regulated by ubiquitylation.
The PRAS40 protein also functions as a negative regulator of the kinase activity of mTOR. PRAS40 inhibits the ability of mTORC1 to undergo autophosphorylation and also inhibits mTORC1 from interacting with several downstream substrates.
mTOR Substrates
Many of the proteins that are known to be substrates for mTOR phosphorylation are involved in either metabolic processes or in the regulation of protein synthesis. As indicated above, Raptor plays an important role in the recruitment of mTOR substrates to mTORC1.
mTORC1 regulates the levels of several transcription factors by altering their transcription and translation. In the context of mTORC1 function in metabolism, one important target of mTORC1 action is HIF1α. Another transcription factor whose levels are affected by the activity of mTORC1 is MYC.
Both mTORC1 and mTORC2 have been shown to phosphorylate several protein kinases including AMPK, PKA, PKC, PKG, AKT, and ribosomal protein S6 kinase 1 (p70S6K; encoded by the RPS6KB1 gene). The p70S6K enzyme, as well as PKC and PKG, belongs to the large AGC family of kinases which includes 10 genes encoding ribosomal protein S6 kinases.
Regulation of protein synthesis by mTORC1 involves the phosphorylation of p70S6K as well as eukaryotic initiation factor 4E binding protein 1 (4EBP1; encoded by the EIF4BP1 gene). mTOR also phosphorylates PRAS40 which results in the de-repression of mTORC1.
As indicated in the Regulation of mTORC1 Activity in the Regulation of Autophagy section below, mTOR phosphorylates several proteins involved in the processes of autophagy including ULK1 and Atg13 resulting in the inhibition of autophagy.
Role of Amino Acids in the Regulation of mTORC1 Activity
With respect to regulation of mTORC1 activity by amino acids the primary regulators are leucine, arginine, glutamine, and serine. The ability of amino acids to regulate mTORC1 activity is coupled to the activity of plasma membrane amino acid transporters. The heterodimeric amino acid transporter, that is composed of the SLC3A2 and SLC7A5 encoded proteins (4F2hc and LAT1 respectively), in conjunction with the SLC1A5 encoded glutamine transporter (ASCT2) transports glutamine out of the cell and leucine into the cells contributing to the activation of mTORC1.
In addition to the role of plasma membrane amino acid transporters in the regulation of mTORC1 activity, intracellular amino acid transporters also contribute to the ability of amino acids to regulate the activity of mTORC1. As indicated, mTORC1 is activated by association with membranes of the lysosomes and amino acid transporters associated with lysosomal membranes contribute to activation of mTORC1. Several transporters of the SLC36 family, the proton-coupled amino acid transporters (PAT) are involved in the activation of lysosome-associated mTORC1. Both PAT1 (encoded by the SLC36A1 gene) and PAT4 (encoded by the SLC36A4 gene) contribute to mTORC1 activation. The role of intracellular amino acid transporters in the regulation of mTORC1 activity has led to them being described as transceptors.
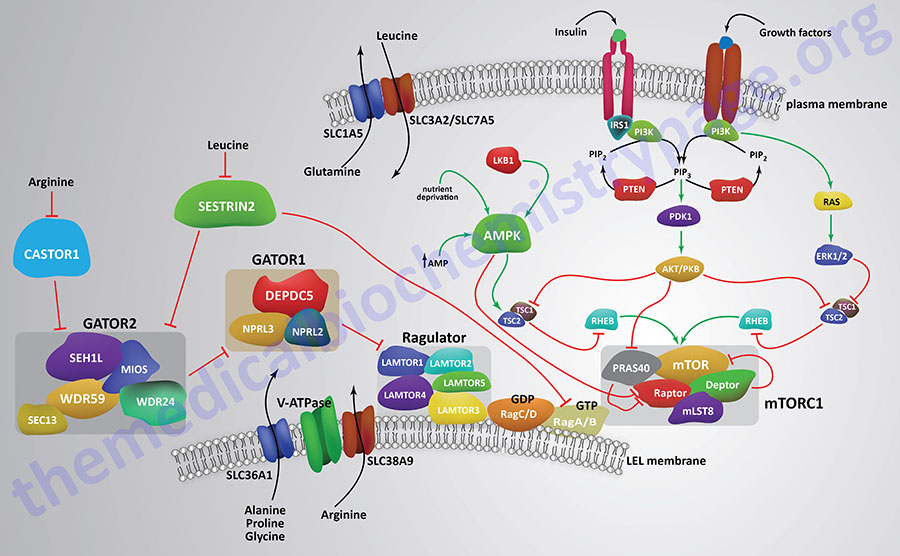
Another amino acid transporter that has been shown to contribute to the activation of the mTORC1 is encoded by the SLC38A9 gene. This transporter is a member of the sodium–coupled neutral amino acid (system N and system A) transporter (SNAT) family. The SLC38A9 encoded transporter is also known as SNAT9. SLC38A9 has been shown to interact with proteins of the Ragulator complex. The Ragulator complex interacts with and modifies the activity of proteins of the Ras-related GTP binding protein (Rag) family. SLC38A9 has been shown to interact with the LEL membrane associated V-ATPase coupling the complex to proton transport. When arginine activates the gating function of SLC38A9 it stimulates GDP release from RagA/B.
The contribution of various amino acid transporters to the activation of mTORC1 represents only a small part of the overall complexity associated with the activation of this complex. The interaction of numerous proteins with the mTORC1 to generate a fully active complex generates what is termed a supercomplex.
Ragulator Complex
The Ragulator complex itself consists of proteins encoded by genes of the LAMTOR (Late endosomal/lysosomal Adaptor, MAPK and mTOR activator) family, of which there are five genes. All five LAMTOR encoded proteins form the pentameric Ragulator complex. The LAMTOR1 protein serves to anchor the Ragulator complex to the LEL membranes. The association of LAMTOR proteins with mTORC1 (as well as mTORC2) provides the scaffold for the binding of AMP-activated protein kinase (AMPK) and mitogen-activated protein kinases (MAPK) facilitating the signaling pathways elicited by these kinases.
The LAMTOR/Ragulator pentameric complex interacts with heterodimeric Rag proteins where the RagA or RagB associate with RagC or RagD. The RagA/B proteins bind GTP whereas the RagC/D proteins bind GDP under conditions of amino acid sufficiency. The Rag heterodimers interact with the RAPTOR protein to facilitate mTORC1 translocation to the LEL membrane. The LAMTOR/Ragulator complex serves as the guanine nucleotide exchange factor (GEF) for the RagA and RagB proteins.
GATOR1 and GATOR2 Complexes
Regulation of the GTPase activity of RagA and RagB is accomplished by a multiprotein GTPase activating (GAP) complex identified as GATOR1 (Gap Activity TOward Rags 1). The GATOR1 complex is composed DEP domain containing 5 (DEPDC5), nitrogen permease regulator 2-like protein (NPRL2), and NPRL3. The ability of RagA to be loaded with GTP is, in part, regulated by a lysosomal membrane-associated GPCR identified as GPR137B.
The RagC and RagD proteins are also regulated by a GTPase activating protein (GAP) complex consisting of the protein folliculin (encoded by the FLCN gene) and the interacting partner proteins identified as folliculin interacting proteins 1 and 2 which are encoding by the FNIP1 and FNIP2 genes, respectively.
The recruitment of GATOR1 to the membranes of the LEL involves the multiprotein complex identified as KICSTOR which gets its name from the genes encoding proteins of the complex: KPTN, ITFG2, C12orf66 (KICS2), and SZT2, and mTOR. The KPTN gene encodes kaptin, actin binding protein. The ITFG2 gene encodes integrin alpha FG-GAP repeat containing 2. The KICS2 gene encodes KICSTOR subunit 2. SZT2 encodes SeiZure Threshold 2 homolog subunit of KICSTOR complex. KICSTOR has been shown to be involved in amino acid and glucose deprivation-mediated inhibition of the mTORC1.
Another protein complex that is identified as GATOR2 has been shown to indirectly activate the mTORC1 through its ability to block the activity of the GATOR1 complex. The GATOR2 complex consists of the proteins Mios, WD repeat-containing protein 24 (WDR24), WDR59, SEH1-like nucleoporin (SEH1L), and SEC13. The SEC proteins are homologues of Drosophila melanogaster secretory mutant genes.
CASTOR1
The amino acid arginine, and most likely leucine as well, has been shown to play an important role in the activation of the mTORC1 via its ability to enhance the activity of the LAMTOR/Ragulator complex as well as through its ability to inhibit the activity of a protein identified as CASTOR1 (Cytosolic Arginine Sensor for mTORC1). CASTOR1 normally inhibits the activity of the GATOR2 complex, thus the inhibition of CASTOR1 by arginine allows GATOR2 to inhibit GATOR1 which in turn would normally inhibit the Rag heterodimers from activating the mTORC1. CASTOR1 can form homodimers or it can heterodimerize with a related protein termed CASTOR2.
Sestrin2
Another protein that inhibits GATOR2 is Sestrin2 which is encoded by the SESN2 gene. Sestrin2 is a member of a family of three proteins that are small stress induced proteins. The ability of Sestrin2 to inhibit GATOR2 is regulated by its interaction with leucine. The interaction of leucine with Sestrin2 leads to inhibition of the ability of Sestrin2 to inhibit GATOR2. The net effect is that this action of leucine results in GATOR2-mediated inhibition of GATOR1 which in turn allows RagA/B to activate the mTORC1.
SAMTOR
Methionine also functions to regulate the activity of mTORC1 in the form of S-adenosylmethionine, SAM (also designated as AdoMet). Methionine is transported into cells primarily via the SLC7A5 and SLC43A2 transporters. The conversion of methionine to SAM is catalyzed methionine adenosyltransferase (MAT).
SAM interacts with a protein that is identified as SAMTOR (S–AdenosylMethionine sensor upstream of mTORC1) which is encoded by the BMT2 (base methyltransferase of 25S rRNA 2 homolog) gene. SAMTOR normally co-localizes with GATOR1 and KICSTOR but when SAM interacts with SAMTOR its ability to interact with GATOR1 is impeded which, in turn, results in decreased inhibition of the Ragulator complex by GATOR1. The net effect of methionine, via SAM, is activation of the mTORC1.
The ability of SAMTOR to interact with SAM is dependent upon the activity of the arginine methyltransferase identified as PRMT1. If there is sufficient methionine, the levels of SAM will be elevated. SAM interacts with SAMTOR inducing its dissociation from the GATOR1 complex. SAM also interacts with PRMT1 allowing the activity of PRMT1 to be directly responsive to changes in the levels of SAM. The dissociation of SAMTOR allows for PRMT1 to interact with the GATOR1 complex. PRMT1 then methylates the NPRL2 protein of the GATOR1 complex which results in inhibition of the GAP activity of GATOR1. The net effect of sufficient methionine, via PRMT1, is activation of the mTORC1.
Regulation of mTORC1 Activity by Ubiquitylation
As detailed in the previous section, the activity of the mTORC1 is tightly regulated by the intracellular levels of several amino acids. When amino acid levels decline the overall level of amino acid charged tRNAs also declines. The decrease in charged tRNAs results in activation of the kinase, GCN2, which is a member of the eIF-2α kinases. The activation of GCN2 results from direct interaction of the kinase with uncharged tRNA. When eIF-2α is phosphorylated there is preferential translation of stress responsive mRNAs such as the mRNA encoding the transcription factor, ATF4 (activating transcription factor 4). One of the targets of ATF4 is the gene encoding Sestrin2.
In addition to phosphorylating eIF-2α, GCN2 phosphorylates the protein identified as F-box only protein 22 (encoded by the FBXO22 gene). The FBXO22 protein is a substrate-recognition component of the SCF (SKP1-CUL1-F-box protein)-type E3 ubiquitin ligase complex. FBXO22 is localized to the nucleus and the cytosol. The FBXO22 containing ubiquitin ligase complex ubiquitylates numerous substrates including several lysine demethylases and several methylated proteins such as p53 and PTEN. When FBXO22 is phosphorylated, it is retained in the cytosol and in this location one of its substrates is mTOR within the mTORC1. When mTOR is ubiquitylated it can no longer phosphorylate its substrates, several of which are involved in the activation of protein synthesis. Thus, under conditions of amino acid deprivation mTORC1 activity is inhibited by ubiquitylation of mTOR and protein synthesis is reduced.
The ubiquitylation of mTOR represents another mechanism by which nutrients, such as amino acids, control the activity of the mTORC1. The major sensors that are involved in the activation of mTOR ubiquitylation are depletion of Arg, Gln, and Leu.
mTOR Activity in the Regulation of Metabolism
Both the mTORC1 and mTORC2 exert direct and indirect effects resulting in alterations in the glycolysis pathway. As indicated above, mTORC1 affects the level of transcription factors such as HIF1α and MYC, both of which are involved in the regulation of expression of several genes responsible for the metabolism of glucose. When mTORC1 is active there is an increase in the level of expression of the gene (HIF1A) encoding HIF1α.
As described in more detail in the Hypoxia and Metabolism page, HIF1α activates the transcription of numerous genes encoding proteins involved in glucose metabolism. These genes encode those that code for pyruvate kinase M (PKM) gene, the phosphofructokinase 1 encoding (PFKL and PFKP) genes, the glucose-6-phosphate isomerase (GPI) gene, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, the phosphoglycerate kinase 1 (PGK1) gene, the phosphoglycerate mutase (PGAM1) gene, the hexokinase 1 (HK1) and HK2 genes, the enolase 1 (ENO1) and ENO2 genes, the fructose-1,6-bisphosphate aldolase (ALDOA) gene, the pyruvate dehydrogenase kinase 1 (PDK1; also designated PDHK1) gene, the SLC2A1 gene (encoding GLUT1), the SLC2A3 gene (encoding GLUT3), the lactate dehydrogenase A (LDHA) gene, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, and the SLC16A3 gene (encoding the lactate transporter, monocarboxylate transporter 4: MCT4).
Activation of mTORC1 also results in increased transcription of the gene (PFKFB3) that encodes the inducible form of the bifunctional enzyme phosphofructokinase-2/fructose-2,6-bisphosphatase (PFK-2/F-2,6-BPase, or commonly just PFK-2). The activation of the PFKFB3 gene is dependent on the activity of HIF1α whose expression is upregulated by mTORC1.
mTORC1 activity also enhances the expression of the PKM2 isoform of pyruvate kinase. As discussed in the Metabolic Alterations Associated with Cancer page the PKM2 isoform is expressed in all cancer cells and in most proliferating cells. The mTORC1-mediated switch to PKM2 synthesis results from the activation of HIF1α and MYC. This activation of PKM2 by HIF1α results in an activation loop since PKM2 serves as a transcriptional co-activator of the HIF1A gene. PKM2 also induces increased mTORC1 activity through its phosphorylation and inhibition of the mTORC1 inhibitor PRAS40.
In the context of pyruvate production, mTORC1 also activates expression of the LDHA and LDHB genes that encode lactate dehydrogenase, LDH. The increased expression of LDH results in increased reduction of pyruvate to lactate. The LDHA gene is also transcriptionally activated by HIF1α. Increased lactate results in acidification of the microenvironment which promotes additional activation of HIF1α.
The mTORC2 also regulates the metabolism of glucose via mechanism that are dependent upon activation of AKT as well as mechanisms that are independent of AKT. mTORC2 activity results in increased levels of hexokinase (HK1 and HK2), PFK2, and the glucose transporter, GLUT1. Through the activation of expression of the MYC transcription factor, mTORC2 exerts effects on glucose metabolism that are similar to those of mTORC1. mTORC2 enhances the phosphorylation of several histone deacetylases (HDAC) rendering these enzymes less capable of deacetylating histones in the control regions of transcription factors of the FOXO family which results in de-repression of MYC gene transcription.
By enhancing glucose metabolism to pyruvate, mTORC1 and mTORC2 promote carbon entry into the TCA cycle. In addition to enhancing substrate entry in the TCA cycle, mTORC1 is responsive to various metabolic intermediates of the cycle. Addition effects on mitochondrial metabolism are exerted by mTORC1 which includes activation of expression of genes encoding protein components of the electron transport chain. One primary target of mTORC1 that results in increased mitochondrial metabolism is PGC-1α.
Not only are mTORC1 and mTORC2 responsive to amino acids, mTORC1 is known to play a role in enhancing the metabolism of glutamine. These effects of mTORC1 are exerted via its activation of MYC which is a transcription factor involved in the repression of the expression of the GLS2 gene that encodes the mitochondria-localized form of glutaminase. mTORC1 also inhibits several FOXO family transcription factors that normally inhibit the expression of the GLUL gene which encodes glutamine synthetase. This effect, in conjunction with MYC, results in enhanced levels of glutaminase. In addition to the GLS2 gene, mTORC1 is involved in the regulated expression of the gene (GLUD1) encoding glutamate dehydrogenase. mTORC1 inhibits activating transcription factor 4 (ATF4; previously identified as CREB2) which normally inhibits the expression of the GLUD1 gene. The overall effects of mTORC1 on glutamine metabolism result in increased utilization of the carbons, via entry into the TCA cycle and as a consequence gluconeogenesis, for energy production.
The role of mTORC1 in lipid homeostasis results in increased lipogenesis. Conversely, when the activity of mTORC1 is inhibited there is an increase in triglyceride breakdown and fatty acid oxidation. The role of mTORC1 in lipogenesis is medicated by activation of expression the SREBP genes. As indicated, mTORC1 phosphorylates and activates p70S6K which in turn phosphorylates and activates SREBP-1c. SREBP-1 activates the expression of several genes involved in lipid synthesis including ACLY (encoding ATP-citrate lyase), ACACA (encoding acetyl-CoA carboxylase 1, ACC1), FASN (encoding fatty acid synthase), and SCD (encoding stearoyl-CoA desaturase).
Adipose tissue metabolism is also regulated by mTORC1 through its regulation of the activity of peroxisome proliferator-activated receptor gamma (PPARγ). PPARγ is a major regulator of adipogenesis. mTORC1activity results in reduced triglyceride breakdown in adipocytes via its inhibition of the activity of adipose triglyceride lipase (ATGL). The repression of ATGL activity by mTORC1 result from enhanced expression of the transcription factor, Egr1 (early growth response 1), that represses the PNPLA2 gene that encodes ATGL.
mTORC1 Activity in the Regulation of Autophagy
Autophagy is suppressed when mTOR is activated by a variety of different growth factor receptors. Upstream signaling proteins that stimulate the activity of mTOR include the Ser/Thr kinases, AKT, phosphatidylinositol-3-kinase (PI3K), and several members of the MAPK family of kinases. AKT was originally identified as the tumor inducing gene in the AKT8 retrovirus found in the AKR strain of mice. AKT is also known as PKB. Humans express three genes in the AKT family identified as AKT1 (PKBα), AKT2 (PKBβ), and AKT3 (PKBγ). Under conditions of energy depletion, such as nutrient starvation, the master metabolic regulatory kinase, AMPK, is activated which phosphorylates and inhibits mTOR, thereby promoting autophagy induction. The tumor suppressor p53 can also activate autophagy under conditions of genotoxic stress through repression of mTOR activity.
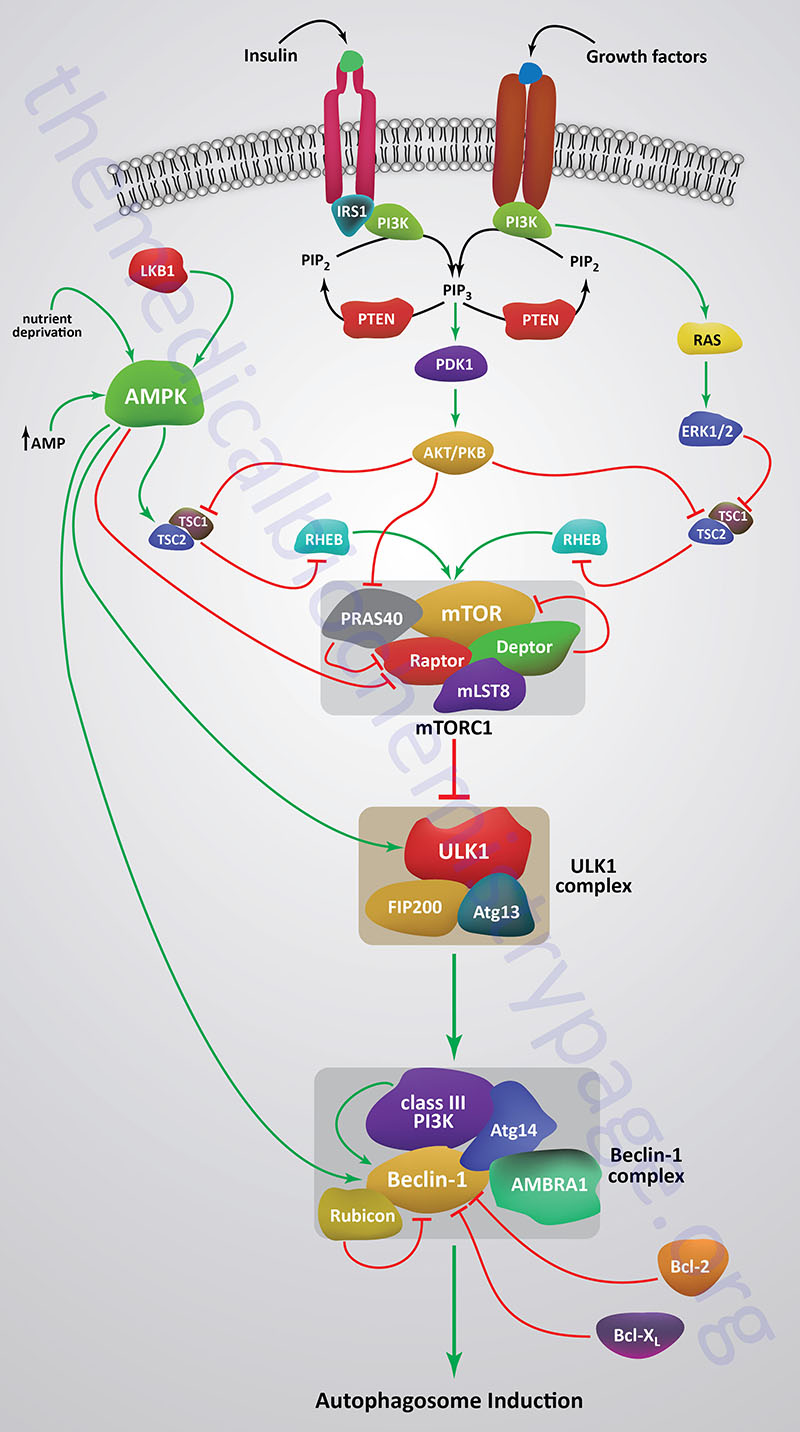
Growth factor receptors that regulate the kinase activity of mTOR activate PI3K which phosphorylates membrane localized phosphatidylinositol-4,5-bisphosphate (PIP2; PtdIns-4,5-P2) generating phosphatidylinositol-3,4,5-trisphosphate (PIP3; PtdIns-3,4,5-P3). PIP3 then activates the kinase PDPK1 (PIP3-dependent protein kinase 1; also known as 3-phosphoinositide dependent protein kinase 1) that then phosphorylates and activates AKT. PDPK1 only partially activates AKT which requires an additional phosphorylation at a Ser (S473) residue catalyzed by the mTORC2.
When AKT is activated it phosphorylates the TSC2 subunit of the tuberous sclerosis complex (TSC). AKT phosphorylates TSC2 on two Ser (S939 and S981) residues and one Thr (T1462) residue. The TSC includes TSC1, TSC2, and TBC1 domain family member 7 (encoded by the TBC1D7 gene). TSC2 functions as a GTPase activating protein (GAP) that inhibits the activity of the G-protein Rheb (Ras homolog expressed in brain). Phosphorylation of TSC2 results in inhibition of the GAP activity which allows Rheb to remain active which ultimately leads to increased mTORC1 activity.
Within the context of autophagy there are two related downstream kinases whose activities are regulated by activated mTORC1. These Ser/Thr kinases are known as UNC-51-like autophagy activating kinases -1, and -2, which are encoded by the ULK1 and ULK2 genes. Humans express a third related gene, UKL3, whose encoded kinase is primarily involved in the regulation of the activity of the sonic hedgehog (SHH) protein. However, evidence does indicate that ULK3 can function in some capacity in the regulation of autophagy.
The UNC genes represent a large family of genes identified in the round worm C. elegans that, when mutated, result in uncoordinated motor activity. The human ULK1 protein functions similarly to the yeast ATG1 protein. ULK1 (or its close family member ULK2) interacts with several other proteins to form a large complex that is required for the formation of an autophagosome. The other proteins that complex with ULK1/ULK2 include ATG13, ATG101, and RB1CC1 (FIP200/ATG17).
As indicated, under conditions of nutrient deprivation AMPK is activated, thereby phosphorylating and inhibiting the kinase activity of mTOR within mTORC1. When the mTORC1 is active it phosphorylates ULK1 on a regulatory Ser residue (S757) thereby, inhibiting the activation of ULK1. Thus, AMPK-mediated inhibition of mTORC1 activity allows ULK1 to induce autophagy. AMPK also directly phosphorylates ULK1 on two different Ser residues (S317 and S777) which results in activation of the autophagy induction properties of ULK1.
In addition to the role of AMPK in the regulation of ULK1/ULK2 activity via its effects on mTORC1 activity, this master metabolic regulatory kinase also regulates the autophagy complex downstream of ULK1/ULK2 at the level of the beclin 1 complex, also called the class III PI3K complex.
The beclin 1 complex consists of several proteins including, but not limited to beclin 1 (the mammalian homolog of yeast ATG6), ATG14 (often referred to as ATG14-like, ATG14L), ultraviolet irradiation resistance-associated gene (UVRAG), AMBRA1 (autophagy and beclin 1 regulator 1), RUBICON (RUN and cysteine-rich domain containing beclin 1 interacting protein), and SH3 domain containing GRB2 like endophilin B1 (encoded by the SH3GLB1 gene; also known as BIF-1).
The primary function of the activation of the beclin 1 complex is to activate the class III PI3K encoded by the PIK3C3 gene. PIK3C3 encodes a p110 catalytic subunit which forms a heterodimer with the p150 regulatory subunit encoded by the PIK3R4 gene. Activation of the class III PI3K results in the production of phosphatidylinositol 3-phosphate (PI3P) at the site of phagophore formation. One important role of PI3P is the recruitment of the WIPI proteins (human homologs of the yeast ATG18 protein) to the phagophore membrane. In addition, PI3P may recruit other downstream effector proteins of the autophagy process such as the ATG5-ATG12-ATG16L1 complex. The function of the beclin 1 protein within the complex can be inhibited via interaction with the pro-survival (anti-apoptotic) proteins BCL-2 and BCL-XL (see Mitochondrial Apoptosis section in the Protein, Organelle, and Cell Turnover page).
BCL-2 and BCL-XL bind to the BH3 domain of beclin 1 thereby inhibiting its activity. The interaction of beclin 1 with either of the two pro-survival proteins can be inhibited by phosphorylation of beclin 1 or BCL-2 or by the ubiquitylation of beclin 1. Therefore, it is clear that the beclin 1 complex plays an important role in promoting mitochondrial apoptosis as well. Mitochondria localized beclin 1.
Regulation of mTORC2 Activity
The mechanisms that regulate the activity of the mTORC2 are less well characterized in comparison to those regulating the mTORC1. Nutrient status and growth factor signal transduction cascades are known to affect the activity of mTORC2. The localization of mTORC2 is predominantly at the inner surface of the plasma membrane but is also found associated with endosomal membrane.
Similar to the role of growth factor activation of PI3K in regulation of mTORC1 activity, PI3K is also involved in the activation of mTORC2 activity. The generation of PIP3 by PI3K results in the tethering of mTORC2 to the plasma membrane and its activation. When mTORC2 is activated by PIP3 it phosphorylates AKT leading, ultimately, to the activation of the mTORC1. Active AKT phosphorylates and inhibits the TSC1/TSC2 complex which normally inhibits mTOR in the mTORC1, Thus, growth factor activation of mTORC2 will lead to activation of mTORC1.
Nutrient status, particularly glucose starvation and glutamine depletion, lead to the activation of mTORC2. The activation of mTORC2 as a consequence of glucose and glutamine deprivation results in the maintenance of the flux of glucose and glutamine into the hexosamine biosynthesis pathway. Nutrient depletion also results in the activation of AMPK which directly phosphorylates mTOR in the mTORC2 just as is the case for the role of AMPK in the regulation of mTORC1 in response to nutrient depletion.
Adrenergic receptor stimulation has also been shown to be involved in the regulation of mTORC2 activity. Within skeletal muscle the activation of β2-adrenergic receptors by epinephrine results in activation of mTORC2. The activation of mTORC2 in the cells promotes glucose uptake and protein synthesis. Stimulation of α1-adrenergic receptors in cardiomyocytes leads to activation of mTORC2 thereby promoting glucose uptake.