Last Updated: October 24, 2025
Introduction to the Wnt Family of Proteins
The Wnt genes constitute a large family of cysteine-rich, secreted glycoproteins that are involved in critical aspects of early embryonic development. The term wnt is an amalgam of wingless (Wg) and int. The Wg gene was identified as a locus in Drosophila required in each segment for the establishment of normal pattern. In the absence of Wg, posterior pattern elements (i.e. naked cuticle) are replaced by mirror image duplications of anterior denticle bands.
The int loci (there were two loci termed int-1 and int-2) were originally identified in mice as sights of frequent integration (hence int loci) of the Moloney murine leukemia virus. Subsequently it was shown that the murine int-1 locus was homologous to Drosophila Wg and the family name was changed to Wnt to reflect both origins.
There are 19 known human WNT genes, as well as the same number of murine genes. The human and mouse Wnt proteins, and their encoding genes, carry the same nomenclature: WNT1, WNT2, WNT2B, WNT3, WNT3A, WNT4, WNT5A, WNT5B, WNT6, WNT7A, WNT7B, WNT8A, WNT8B, WNT9A, WNT9B, WNT10A, WNT10B, WNT11, WNT16. The expression patterns of several of the mouse Wnt genes are described in the Table below.
Table of Wnt Expression Patterns in Mice
| Gene | Embryonic Expression | Adult Expression |
| Wnt1 | brain, spinal cord | testes |
| Wnt2 | ventral-lateral mesoderm, heart, allantois | lung, brain, heart |
| Wnt3 | brain, spinal cord, limbs | thalamus, Purkinje cells |
| Wnt3A | brain, primitive streak | lung |
| Wnt4 | not determined | brain, lung |
| Wnt5A | ventral brain, spinal cord | heart, lung |
| Wnt5B | not restricted | heart, brain, liver, lung |
| Wnt6 | not determined | testes |
| Wnt7A | not determined | brain, lung |
| Wnt8 | not determined | brain |
Classification of Wnt Activities
Results from experiments designed to ectopically express Wnt proteins in mammary epithelial cells and in Xenopus embryos allowed for the loose division of the Wnt proteins into two classes. One class are the axis inducing and transforming Wnt1 class. All the Wnt proteins of this class are capable of transforming mammalian cells in culture and of inducing a secondary neural axis when ectopically expressed in the future ventral side of early frog embryos. The Wnts in this first class are the mouse Wnt1, Wnt3A and Wnt7A genes and the Xenopus Xwnt1, Wnt3A, Wnt8 and Wnt8B genes.
The second class of Wnt proteins is the non-transforming Wnt5A class. This class of Wnt proteins cannot induce the transformed state in cells in culture and do not induce a secondary neural axis in frog embryos. The Wnt proteins of this class include the mouse Wnt4 and Wnt5A genes and the Xenopus Xwnt5A and XWnt4 genes. Chimeric proteins generated by combining elements of Xwnt8 (a Wnt1 class protein) and Xwnt5A (a Wnt5A class protein) and then used for axis induction experiments in frog embryos, demonstrated that the different activities of the two Wnt classes is determined by the C-terminal portion of the proteins.
The Wnt proteins are capable of inducing cells to proliferate, to differentiate, and to survive by signaling through both autocrine and paracrine pathways. The Wnt proteins exert these effects on cells and during early development by interacting with specific cell-surface receptors which in turn initiate a signaling cascade culminating with changes in gene expression. It is this Wnt-induced alteration in the expression of specific genes that leads to the multitude of effects observed for Wnt proteins.
Wnt Signal Transduction Pathways
Frizzled Receptors
Wnts are soluble secreted factors that exert the effects described above through interaction with cell surface receptors. Wnt proteins bind to receptors of the Frizzled family, named after the Drosophila tissue polarity gene, frizzled. Humans express 10 genes (FZD1-FZD10) encoding Frizzled proteins.
Structurally the Frizzled receptors have an extracellular Wnt-binding domain, seven transmembrane-spanning regions, and an intracellular C-terminal tail. The presence of the seven transmembrane-spanning domains places the Frizzled proteins in the superfamily of receptors known as the G-protein coupled receptors, GPCR (see the Signal Transduction Pathways: G-Proteins and GPCR page for details of this receptor family).
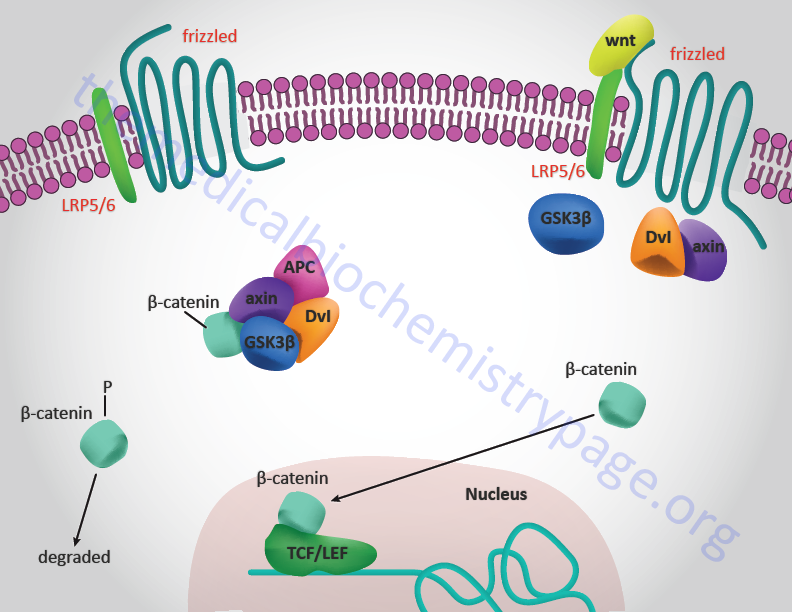
In addition to the frizzled protein, there are co-receptors of Wnt binding that are members of the low-density lipoprotein (LDL) receptor-related proteins (LRP, of which there are at least eleven members). The first Wnt co-receptor identified was the Drosophila gene arrow which showed homology to mammalian LRP5 and LRP6. Thus, the Wnt co-receptor is identified as LRP5/6. The interactions between Wnt, Frizzled, and LRP5/6 is referred to as the canonical Wnt signaling pathway.
Additional components of the canonical Frizzled receptor-Wnt interaction pathway are the soluble Wnt inhibitors which consist of secreted proteins containing a cysteine-rich domain (CRD) similar to that in the ligand binding domain of the Frizzled receptors. This class of Wnt inhibitory proteins are termed frizzled receptor-like proteins (FRP). By binding Wnts, FRP proteins can sequester Wnts away from the cell-surface Frizzled receptors and thereby, reduce the effective concentration of available Wnt protein.
Direct evidence for the interaction of Wnts with Frizzled receptors came from several studies. When Drosophila frizzled-2 (Dfz2) was expressed in fly cells in culture, the cells attained the ability to bind the Wg protein. When rat Frizzled 1 (RFz1) or human Frizzled 5 (Hfz5; encoded by the FZD5 gene) were expressed in frog embryos, Xwnt5A and Xwnt8 activated axis duplication through interaction with these exogenous receptors. The presence of a large family of Frizzled receptors suggests that there may exist the ability to differentially bind various Wnt proteins. Indeed, evidence shows that Dfz1, Dfz2, MFz4 (mouse Frizzled 4), MFz7, MFz8 and HFz5 but not MFz3 were able to bind Wg in fly cell in culture. Since Wg can bind to at least six different Frizzled receptors it is likely that multiple Wnts can be bound by any given Frizzled receptor and that many Wnts will be able to bind to multiple Frizzled receptors. Since endogenous Wnt expression patterns overlap (particularly in the mouse hindbrain) there may be functional redundancy in Wnt expression.
It is also probable that overlapping patterns of Wnt expression may allow for different Wnts to antagonize each other during development. Evidence for this latter possibility has been most clearly demonstrated in studies of axis duplication in Xenopus. Prior expression of Xwnt-5a blocks the axis inducing activity of Xwnt8. The precise mechanism for this interference is not clear. It could be that Xwnt5A blocked Xwnt8 action by receptor competition or by blocking the action of intracellular signaling components.
Since the Frizzled receptors have no enzymatic motifs on their intracellular domains, the Wnt signals are likely transmitted through receptor coupling to intracellular proteins that are functionally related to other members of the G-protein family. However, no distinct GTP-dependent activity has been shown to be coupled to the Wnt signal transduction pathways.
Dishevelled
One candidate family of proteins, shown by genetic studies in Drosophila to be downstream of Wg, is that characterized by dishevelled (Dvl or Dsh). Members of the dishevelled family encode cytoplasmic proteins with no known enzymatic functions. Three homologues have been identified in humans (DVL1, DVL2, and DVL3) and shown to have overlapping patterns of expression. This overlapping pattern of expression suggests redundancy of function for Dishevelled proteins. This supposition is supported by the fact that targeted deletion of the mouse Dvl-1 gene results in mice that are structurally normal.
There are three regions of sequence homology between the different dishevelled family members. The N-terminally conserved domain is termed the DVL homology domain. This domain is similar to the C-terminus of Axin (encoded by the AXIN1 gene in humans and the D. melanogaster fused gene) a protein shown to interfere with the formation of the endogenous dorsal axis and to prevent Xwnt8 from inducing an ectopic axis.
The central portion of dishevelled proteins contains a PDZ domain. The PDZ domain is composed of 80-90 amino-acids which was originally identified in the three proteins, Post synaptic density protein (PSD95), D. melanogaster Disc large tumor suppressor (Dlg1), and Zonula occludens-1 protein (zo-1). The PDZ domain is present in many other proteins and has been shown to bind C-terminal peptides of the sequence motif X-(S/T)-X-V (where X is any amino acid). Proteins containing PDZ domains are involved in the anchoring of membrane receptor proteins to cytoskeletal components. The D. melanogaster Dvl PDZ domains have been shown to be essential for downstream signaling in frog embryos.
The third homology domain in dishevelled proteins is the DEP domain. The DEP domain is a globular domain consisting of around 80 amino acids whose name is derived from the three proteins in which it was originally identified (Dishevelled, EGL-10, and Pleckstrin). In humans the EGL-10 homolog is encoded by the RGS7 (regulator of G-protein signaling 7) gene. The DEP domain has similarity to GTPase activating proteins (GAP) and guanine nucleotide exchange factors (GEF). The DEP domain may, therefore, be important for recruiting downstream targets in G-protein-coupled pathways.
Disheveled provides the scaffold for the binding of Axin and glycogen synthase kinase 3β (GSK3β) to the activated FZD-LRP5/6 complex. This recruitment results in the inhibition of GSK3β by dishevelled. GSK3β is a predominantly cytoplasmic kinase originally identified for its role in glycogen metabolism. GSK3β has been shown to be the mammalian homolog of the D. melanogaster gene zeste-white-3 (zw3). Genetic studies showed that zw3 functioned downstream of Dvl and probably inactivated Wg signaling. The mammalian GSK3β protein can partially compensate for loss of zw3 during D. melanogaster embryogenesis.
The fact that activation of dishevelled leads to an inactivation of GSK3β activity was demonstrated by the failure of GSK3β to phosphorylate the microtubule-associated protein tau in Dvl-1 overexpression studies. When a dominant negative form of Xenopus GSK3β (dnGSK3β), which lacks the kinase domain, is expressed in Xenopus embryos an ectopic axis forms which closely resembles the axis induced by ectopic expression of Xwnt-8 and Xdsh. In contrast, wild-type XGSK3β is able to suppress the ectopic axis inducing activities of Xwnt-8 and Xdsh. This suggests that the normal function of GSK3β is to suppress Wnt signaling from Dvl.
β-Catenin
The consequences of Wnt-induced inhibition of GSK3β activity is that the next protein identified in the cascade, β-catenin, is stabilized. β-Catenin is a member of a multigene family of proteins characterized by the presence of a domain originally found in the D. melanogaster Armadillo (Arm) gene. This family of proteins is termed the Armadillo repeat-containing protein (ARMC) family. Humans express 43 genes in the ARMC family. The ARMC proteins possess tandem repeats with a length of approximately 42 amino acids that serve as binding platform for many proteins. The ARMC family of proteins is just one of seven subfamilies of the Armadillo-like helical domain containing (ARMH) superfamily that consists of 244 genes.
Human β-catenin is encoded by the CTNNB1 gene. The CTNNB1 gene is located on chromosome 3p22.1 and is composed of 21 exons that generate four alternatively spliced mRNAs, three of which encode isoform 1 (781 amino acids) and one that encodes isoform 2 (774 amino acids).
Genetic studies showed that Arm functions downstream of zw3/GSK3β. Evidence indicates that Wnt inhibition of GSK3β activity prevents the turnover of β-catenin leading to its accumulation. The loss of zw3 function or the use of dnGSK3β stabilizes β-catenin and Arm levels in both Drosophila and Xenopus indicating that GSK3β normally functions to lower β-catenin levels. Additionally, the loss of zw3 or the use of dnGSK3β leads to a reduction in the phosphorylation of Arm and β-catenin. These observations indicate that phosphorylation of β-catenin leads to its degradation, by an as yet unidentified mechanism.
There are several structural domains in the β-catenins. The N-terminus is the site of GSK3β-mediated phosphorylation. Another region binds to α-catenin, a cytoplasmic protein with similarity to vinculin which links actin filaments to the adherins junctions. The central region of the protein contains a positively charged groove that is suspected to be involved in interactions with acidic regions in the adenomatous polyposis coli (APC) locus encoded protein, the transcription factor TCF/LEF-1 (T-cell factor/lymphocyte enhancer factor-1) and the cadherin family of calcium-dependent cell-adhesion molecules. The APC gene is a tumor-suppressor linked to inherited and sporadic forms of colon cancer (see the Tumor Suppressor Genes and Cancer page for more information on the connection between APC and Wnts). Mutational analyses have shown that regions of β-catenin that are required for Wnt signaling are separable from those required for cell adhesion suggesting that cadherins are not required for Wnt signaling. However, changes in cadherin expression can affect Wnt signaling and conversely, Wnt signaling can affect cell adhesion. This could possibly lead to altered levels of free β-catenin and thus indirectly alter the level of β-catenin-TCF complexes.
It is not clear whether APC is directly involved in the Wnt signal cascade. However, β-catenin has been shown to directly interact with APC and mutations in APC that led to colon cancer result in a protein that does not bind to β-catenin. Complexes of APC and β-catenin bind to GSK3β. These results suggest that APC functions as a molecular scaffold coordinating the activities of β-catenin and GSK3β such that APC may assist in the ability of GSK3β to phosphorylate β-catenin leading to its degradation. Lending confusion to the role of APC in Wnt signaling is the observation that overexpression of APC in frog embryos positively induced axis duplication instead of suppressing it as expected from the supposition that APC enhances GSK3β activity.
The final link in the Wnt signal cascade is the interaction between β-catenin and the transcription factors of the TCF/LEF-1 family. Proteins of the TCF family bind to DNA and contain a high-mobility group (HMG) domain. There are four TCFs in mammalian cells. They are identified as TCF7 (also known as TCF-1), LEF-1, TCF7L1 (also known as TCF-3), and TCF7L2 (also known as TCF-4). There are three isoforms of the Xenopus XTCF3 gene. The pangolin gene in D. melanogaster and the pop-1 gene of C. elegans are family members. The N-terminus of TCF mediates the interaction with β-catenin. In the absence of β-catenin, TCFs are bound to promoters in Wnt-target genes and repress their expression. Several candidate TCF:β-catenin target genes have thus far been identified. A few are the Xenopus homeobox genes siamois and twin, the secreted protein nodal related 3 (NR3) and fibronectin. The promoters of these genes all contain TCF/LEF binding sites. For a more complete updated listing, visit the Wnt Web site maintained by Roel Nusse which has a table of Wnt/β-catenin targets.
When Wnts bind to the Frizzled-LRP5/6 receptor complex axin, Dvl, and GSK3β are recruited to the complex. This prevents axin from interacting with APC thus, the destructive complex that is comprised of APC, axin, GSK3β and β-catenin (as well as several other proteins) does not form. The role of this latter complex is to induce the phosphorylation of β-catenin which results in its destruction. Thus, when Wnt binds its receptor, β-catenin is released from the destructive complex, is not phosphorylated and can migrate to the nucleus where it interacts with TCF/LEF resulting in transcriptional activation of Wnt target genes.
Control of Wnt Signaling
Control of Wnt signaling occurs at several locations. There are several members of a family of proteins that compete with Wnt binding to frizzled. These proteins are termed soluble frizzled related proteins (sFRP) and they bind Wnt in the extracellular environment preventing Wnt from binding to Frizzled-LRP5/6 receptor complexes. Humans express five genes in the sFRP family.
Another secreted protein that interferes with Wnt interaction with Frizzled-LRP5/6 receptor complexes is Wnt inhibitory factor-1 (WIF1).
An additional inhibitory action is exerted by members of the dickkopf (Dkk) family of proteins, of which there are four members in humans encoded by the DKK1-DKK4 genes. Dickkopf was the name given to a Wnt inhibitor that was first identified in an assay of Xenopus axis formation and is the German word for “big head” or “stubborn”.
Another family of Wnt signaling inhibitory proteins is the naked cuticle (NKD) family of proteins that bind to and inhibit the actions of dishevelled proteins. Humans express two genes encoding NKD family proteins, NKD1 and NKD2. The original naked cuticle (nkd) gene was a segment polarity gene identified in mutants of D. melanogaster.
Wnt Genes and Central Nervous System Development
By using the technique of in situ hybridization, several studies have suggested that Wnt signaling plays an important role during the early patterning of the neural tube. In mouse embryos, the timing of Wnt gene expression in the central nervous system (CNS) indicates that these genes likely assist in the regional specification of the neural tube, in particular in the forebrain region and the spinal cord.
Wnt gene expression within the developing spinal cord exhibits particular patterns of restriction along the dorsal-ventral axis prior to the differentiation of neurons. Wnt1, Wnt3 and Wnt3A are the earliest markers observed to be expressed along the dorsal midline of the developing spinal cord. Wnt4 expression correlates with the region of the presumptive alar plate that is destined to produce sensory interneurons. Wnt7A and Wnt7B are expressed in the ventral spinal cord. Additionally, Wnt-4 is expressed in the ventral region of the floor plate. Since it has been shown that signals emanate from the notochord to induce the formation of the floor plate within the CNS (these are in the form of the inducing protein, sonic hedgehog, shh), the expression patterns of these Wnts suggest that they may play important roles in reinforcing the dorsal-ventral polarities of the CNS established by inducers such as shh. Over-expression of Wnt1 within the spinal cord severely disrupts normal spinal cord morphology, most probably as a result of over proliferation. However, this effect does not interfere with the primary dorsal-ventral patterning of the spinal cord indicating that Wnts likely regulate the patterns of cell proliferation within the spinal cord not the actual patterns induced by other genes.
Several Wnts have been shown to be expressed in the developing forebrain. Wnt5A and Wnt7A are expressed in overlapping domains in the ventral and lateral diencephalon. Wnt1, Wnt3, Wnt3A and Wnt4 are expressed in the dorsal diencephalon. Wnt7B is expressed throughout much of the diencephalon and the optic stalks. Wnt3B and Wnt7B expression extends into the telencephalon along the dorsal regions. These patterns of expression indicate a role for the Wnts as important regulators of the developing forebrain. The pattern of segmentation in the vertebrate hindbrain is divided into regions identified as rhombomeres. Evidence on the expression of numerous genes suggests that the forebrain may be similarly identifiable by specific patterns of segmentation. Wnt gene expression patterns generally coincide with these predicted segments of the forebrain. As an example, an early patch of Wnt3A expression in the diencephalon appears to correspond to the presumptive pretectum (synencephalon). A domain of Wnt3 expression correlates with the future dorsal thalamus. Expression of Wnts in the developing embryonic CNS clearly demarcates dorsal-ventral zones whose subdivisions are comparable to the roof, alar and basal plates of the more posterior neural tube.
Wnt Genes and Embryonic Axis Specification
As indicated above the Wnts are divided into two classes determined in part on an ability, or lack thereof, to induce a secondary neural axis when ectopically expressed in the future dorsal side of frog embryos. The earliest experiments with Wnts and axis duplication were performed with Wnt1. Subsequently, numerous Xenopus Wnt genes (identified as Xwnts) have been isolated and characterized. Three possible functions for Wnts in the establishment of axial pattern have been suggested.
One mechanism suggests that Wnts could assist in the initial establishment of dorsal versus ventral sides of the embryo very early following fertilization and be components of the dorsalizing or ventralizing centers of mesoderm induction. Induction of mesoderm has been established as being critically important in future neuralization of the embryo. A second mechanism has the Wnts acting as parts of the dorsal or ventral signals that modify the inherent character of pre-existing mesoderm. Lastly, Wnts could function by assisting in the production of specified mesoderm, as the primary axis information is established during gastrulation. Strong evidence more closely links Wnts to the latter two models.
Both Xwnt8 and Xwnt11 have the capacity to modify the character of mesoderm that has been induced by other signals. Xwnt8 has the capability to dorsalize regions of the embryo when ectopically expressed. However, it is normally expressed on the ventral side of the embryo during the blastula and gastrula stages. When Xwnt8 is expressed dorsally it has the capability to ventralize the embryo indicating that it is a good candidate for a ventralizing signal. Conversely, Xwnt11 is capable of dorsalizing previously induced mesoderm and the RNA is localized to the marginal zone (future mesodermal tissue is derived from this region of the embryo) with highest levels being found dorsally. When embryos are exposed to uv-irradiation on the vegetal hemisphere they are completely ventralized and form no dorsal structures. Xwnt11 is capable of partially rescuing the dorsal axis in uv-ventralized embryos. Both Xwnt8 and Xwnt11 are incapable of inducing mesoderm on their own, therefore, the above described results indicate that both Wnts must play a role in the modification of pre-existing mesoderm.
Wnt proteins are important mediators of axis establishment when the primary patterns of the embryo are determined during gastrulation. Direct evidence for this has been shown in mice in which Wnt genes have been knocked out by targeted disruption. Expression of Wnt3A is first detected throughout the primitive streak of the mouse embryo in regions fated to generate dorsal mesoderm. Targeted disruption of the mouse Wnt3A gene results in a severe truncation of the body axis posterior to the forelimbs. The most affected tissue being dorsal mesoderm; the notochord is disrupted and the tailbud fails to form. These data demonstrate that Wnt-3a is required for the production of somitic mesoderm and for the generation of all new embryonic mesoderm by the late primitive streak stage.
The Wnt5A and Wnt5B genes are also expressed in the primitive streak during gastrulation in the mouse. Overexpression of Xwnt-5a in frog embryos leads to complex head and tail malformations. Xwnt5A does not ectopically induce mesoderm or change the type of pre-existing mesoderm but can inhibit the normal morphogenetic movements that occur during gastrulation. The chicken Cwnt8C gene (this is the chicken ortholog of the Xenopus Wnt8A gene) is expressed in the posterior marginal zone during gastrulation and later in the primitive streak and Hensen’s node. When Cwnt8C is ectopically expressed in frog embryos it can dorsalize mesoderm and induce axis duplication.
Although no Wnts are candidates for endogenous mesoderm induction or initial patterning, it is clear that this family of proteins is capable of modifying pre-existing mesoderm (Xwnt8, Xwnt11 and Cwnt8C) and affecting gastrulation (Wnt3A, Wnt5A, Wnt5B and Cwnt8C).
Wnt Signaling and Diabetes
Although much of the research that has led to a detailed understanding of the signal transduction pathways initiated by Wnts was carried out in models of early development, evidence has been accumulating demonstrating a significant role for the Wnts in the control of metabolism. In particular Wnt action has been shown to be involved in metabolic control via its actions in both the gut and pancreas. In addition, Wnt signaling has been shown to interact with signaling pathways induced by insulin.
In the gut Wnt has been shown to be involved in regulated expression of the proglucagon gene (gene symbol: GCG). In intestinal enteroendocrine L cells the expression of the GCG gene results in the production of the incretin hormone glucagon-like peptide-1, GLP-1. Incretins are hormones that induce pancreatic insulin secretion in response to food intake. In the pancreas expression of the GCG gene leads to production of glucagon. GLP-1 exerts its effects on the gut, the pancreas and in the brain. In the gut its effects lead to a reduced rate of gastric acid secretion and reduced gastric emptying. In the pancreas GLP-1 induced β-cell proliferation and inhibition of β-cell apoptosis. In the brain GLP-1 actins result in increased satiety leading to reduced desire for food intake.
The GCG gene promoter region contains an enhancer that harbors a canonical Wnt response element that binds TCF factors, in particular the TCF7L2 protein. Genome wide screens for polymorphisms associated with type 2 diabetes demonstrated that two single nucleotide polymorphisms (SNPs) in the TCF7L2 gene were the most frequently occurring SNPs associated with this disease. The significance of Wnt in the control of GLP-1 production was demonstrated by the fact that reduction/loss of either β-catenin or TCF7L2 function completely blocks insulin-stimulated expression of the intestinal GCG gene. In addition, the effects of GLP-1 on the pancreas (i.e. proliferation and anti-apoptosis) are effected via the actions of β-catenin and TCF7L2. In the pancreas insulin inhibits expression from the GCG gene leading to reduced production of glucagon. This action has physiological significance because glucagon is the major counter-regulatory hormone to insulin action. The important role of TCF7L2 in pancreatic function can be demonstrated in experiments that lead to reduction in the levels of TCF7L2. In these types of experiments there is an increased rate of pancreatic β-cell apoptosis, reduced β-cell proliferation, and reduced glucose-dependent insulin secretion.
The demonstration of cross-talk between the Wnt and insulin signaling pathways is important as these observations may eventually lead to novel approaches to the treatment of type 2 diabetes.
TGF-β and BMP Family Members
TGF-β was originally characterized as a protein (secreted from a tumor cell line) that was capable of inducing a transformed phenotype in non-neoplastic cells in culture. This effect was reversible, as demonstrated by the reversion of the cells to a normal phenotype, following removal of the TGF-β. Subsequently, many proteins homologous to TGF-β have been identified. The three closest relatives in the human TGF-β family are TGF-β1 (the original TGF-β) TGF-β2, and TGF-β3. All three of these proteins share extensive regions of similarity in their amino acid sequences. Many other proteins, possessing distinct biological functions, have stretches of amino-acid homology to the TGF-β family of proteins, particularly the C-terminal region of these proteins where a conserved series of six cysteine residues is found. There are at least 42 members of the TGF-β family in humans.
The TGF-β-related superfamily of proteins includes the activin and inhibin proteins. There are activin A, B and AB proteins, as well as an inhibin A and inhibin B protein. The anti-Mullerian hormone (AMH), also known as Mullerian inhibiting substance (MIS), is also a TGF-β-related protein. In addition, members of the bone morphogenetic protein (BMP) family of growth-regulatory factors, first characterized as having effects on bone but known to exert numerous effects in a variety of different systems, are also TGF-β superfamily members. Two additional groups of related proteins belonging to the TGF-β family are the nodal proteins and the growth and differentiation factor (GDF) proteins. Based upon sequence similarities of the various members of the TGF-β superfamily, the superfamily can be divided into four major subfamilies: the TGF-β subfamily, the activin/inhibin subfamily, the decapentaplegic-Vg related (DVR) subfamily which includes the BMP (eleven genes) and GDF (nine genes) proteins, and the fourth subfamily which includes the various divergent family members.
TGFs-β have proliferative effects on many mesenchymal and epithelial cell types. Under certain conditions TGFs-β will demonstrate anti-proliferative effects on endothelial cells, macrophages, and T- and B-lymphocytes. Such effects include decreasing the secretion of immunoglobulin and suppressing hematopoiesis, myogenesis, adipogenesis and adrenal steroidogenesis. Several members of the TGF-β family are potent inducers of mesodermal differentiation in early embryos, in particular TGF-β and activin A.
The BMP family of proteins consists of eleven members in the human genome. Of these eleven BMP proteins, six (BMP2 through BMP7) belong to the TGF-β superfamily.
TGF-β Family Receptors
The TGF-β family members interact with their cell-surface receptors in a dimeric form. There are several classes of cell-surface receptors that bind different TGF-β family proteins with differing affinities. There are also cell-type specific differences in receptor sub-types. Unlike the EGF, PDGF and FGF receptors, the TGF-β family of receptors all have intrinsic serine/threonine kinase activity and, therefore, induce distinct cascades of signal transduction. The TGF-β family receptors consist of a type I and a type II protein component. Currently there are seven type I receptors and five type II receptors known to exist in the human genome.
The seven human type I TGF-β family receptors are identified as TGFBR1 (also known as ALK5), ACVR1 (also known as ALK2), ACVR1B (also known as ALK4), ACVR1C (also known as ALK7), ACVRL1 (also known as ALK1), BMPR1A (also known as ALK3), and BMPR1B (also known as ALK6). The designations ACVR1, ACVR1B, and ACVR1C refer to the genes encoding the activin A receptor type-1, type 1B, and type 1C, respectively. ACVRL1 is the gene encoding the activin A receptor-like type 1 protein. The designations BMPR1A and BMPR1B refer to the genes encoding the bone morphogenetic protein (BMP) receptor type 1A and the BMP receptor type 1B, respectively. The ALK designations were originally used to designate the activin receptors. The five type II TGF-β receptors are identified as TGFBR2, BMPR2, ACVR2, ACVR2B, and AMHR2. The BMPR and ACVR designations for the type II receptors are the same as the similar designations for the type I receptors. The designation AMHR2 refers to the gene encoding the anti-Mullerian hormone receptor type 2.
Ligand binding brings together the type I and type II components of the receptor. This interaction induces the type II receptor to phosphorylate the type I receptor kinase domain. Activation of the type I kinase domain propagates the signal through phosphorylation of the Smad proteins (discussed below). The type I receptors contain a sequence motif: SGSGSG (termed the GS domain), that is the target for type II receptor phosphorylation. Adding to the potential complexity is the fact that specific ligand traps have been identified. These traps are secreted proteins that bind TGF-β ligands in the circulation and thus, interfere with the ability of ligand to dock with receptor.
Table of Ligand-Receptor-Smad Relationships
| Ligands | Receptors | Ligand Traps | Accessory Receptors | Smads |
| TGF-β1 TGF-β2 TGF-β3 | TGFBR2 and TGFBR1 | Latency-associated polypeptide (LAP) Decorin α2-macroglobulin | Betaglycan | Smad2 Smad3 Smad4 |
| Activin proteins | ACVR2 and ACVR1B | follistatin | Smad2 Smad3 Smad4 | |
| Nodal proteins | ACVR2B and ACVR1C | DAN Cerberus | Cripto | Smad2 Smad3 Smad4 |
| BMP2 BMP4 | BMPR2 and BMPR1A | DAN Cerberus Chordin/SOG Follistatin Noggin | Smad1 Smad5 Smad8 Smad4 | |
| GDF5 GDF6 (BMP13) GDF7 (BMP12) | AVCR2B and ACVR1 | Chordin/SOG Follistatin Noggin | Smad1 Smad5 Smad8 Smad4 | |
| BMP7 | ACVR2 and BMPR1A | Chordin/SOG Follistatin Noggin | Smad1 Smad5 Smad8 Smad4 | |
| AMH | AMHR2 and BMPR1B | Smad1 Smad5 Smad8 Smad4 |
Ligand-receptor interactions within the TGF-β family are characterized by two distinct modes. The BMP subfamily of ligands has highest affinity for the type I receptor and low affinity for the type II receptor. The TGF-β subfamily has highest affinity for the type II receptor and low affinity for the type I receptor. The ability of TGF-β ligands to bind and activate receptors is tightly regulated. A group of soluble secreted proteins serves as ligand binding traps that sequester ligand preventing interaction with receptor. One ligand trap comprises the latency-associated polypeptide (LAP) which is actually the pro-region of the TGF-β protein that remains noncovalently bound to the active domain. Other traps include the glycoprotein decorin and α2-macroglobulin which both bind to free TGF-β. Follistatin traps activins and BMPs. Then there are the Noggin, Chordin/SOG (short gastrulation), and DAN/Cerberus families of traps that bind to the BMPs. Another member of the inhibitory class of proteins is BAMBI (BMP and Activin receptors Membrane Bound Inhibitor). BAMBI functions as a decoy receptor whose extracellular domain has sequence similarities to the type I receptors.
In addition to ligand traps that serve to regulate the action of the TGF-β proteins, there exist membrane anchored proteins that serve as accessory receptors or co-receptors. The membrane anchored proteoglycan, betaglycan, has long been known as the TGF-β type III receptor. Betaglycan does not bind to the activins or the BMPs but does interact with inhibins facilitating interaction with the activin receptors. The betaglycan-related protein endoglin facilitates binding of TGF-β family members to the type I receptor ALK1 on endothelial cells. Cripto is a member of the CFC-EGF family of both secreted and membrane anchored proteins. Cripto mediates the binding of Nodal, Vg1, and GDF1 to the activin receptors.
Regulation of TGF-β signaling, at the level of the receptors, also occurs intracellularly. The immunophilin, FKBP12 (a protein that binds to the immunosuppressive drug FK506) binds to the unphosphorylated GS domain of type I receptors preventing ligand-independent phosphorylation and activation of the type I receptor. Following activation, the receptors can be negatively regulated by the inhibitory Smad (I-Smad) Smad7. Smad7 binding leads to the ubiquitylation and degradation of the receptors. This latter process involves the Smad ubiquitylation regulatory factors (Smurfs) which are ubiquitin ligases.
The Role of Smad Proteins in TGF-β and BMP Signaling
The first intracellular mediator of TGF-β signaling was identified in the fruitfly Drosophila melanogaster and the locus was called mothers against decapentaplegic (MAD), decapetaplegic (Dpp) being the Drosophila homolog of the TGF-β protein. Subsequently, related proteins were found in other invertebrates (the round worm genes were called sma) and in vertebrates. Using an amalgam of the worm and fly nomenclature the proteins were named Smad proteins.
There are three classes of Smads: the R-Smads (recepor regulated: are activated as a consequence of phosphorylation by the kinase domain of the type I TGF-β receptors), the I-Smads (inhibitory), and the Co-Smads (common mediator). The R-Smads are Smads 1, 2, 3, 5 and 8. Smad2 and Smad3 respond to signaling via the TGF-β family of receptors, whereas Smad1, 5, and 8 respond to signaling from the BMP family of receptors. The Co-Smad is Smad4. The I-Smad is Smad7.
The R-Smads and Smad4 (co-Smad) contain two conserved structural domains termed MH1 (MAD Homology domain 1) and MH2. The MH1 domain is located at the N-terminus of the Smads and the MH2 domain at the C-terminus. Once phosphorylated, the R-Smads associate with Smad4 and the complex migrates to the nucleus where target gene transcription is affected. It is the MH1 domain of the R-Smads that exhibits DNA-binding activity. The MH2 domain, which is highly conserved among all Smad proteins, is responsible for receptor interaction.
As discussed above, when ligand binds to a type II TGF-β receptor, heterotetrameric complexes are formed with specific type I TGF-β receptors. The formation of these complexes activates the kinase domain of the type I receptors leading to phosphorylation and activation of the associated R-Smad. Phosphorylation of an R-Smad results in association with Smad4 (the Co-Smad). Receptor-mediated phosphorylation of R-Smads occurs on Ser residues in the extreme C-terminal part of the MH2 domain, referred to as Smad tail phosphorylation. Additional sites of Ser and Thr phosphorylation are located within the linker region between the MH1 and MH2 domains. These linker phosphorylation sites are phosphorylated by proline-directed Ser/Thr kinases and this process is referred to as Smad linker phosphorylation. Proline-directed Ser/Thr kinases include members of the MAK kinase family, glycogen synthase kinase 3 (GSK3), and cyclin-dependent protein kinase 5 (CDK5).
Stimulation of cells by TGF-β leads to both positive and negative regulation changes in the expression of several hundred different genes. Both the activation and repression events use the same set of activated Smad proteins. Smad access to target genes along with the recruitment of the various transcriptional co-activators and co-repressors necessary to effect the proper signals in response to TGF-β is dependent upon cell-type specific interactions. Smad proteins thus, cooperate with each other as well as with other DNA binding proteins to ultimately elicit the correct responses. Many different families of DNA-binding proteins have been shown to function as Smad partners. These include homeobox proteins, forkhead proteins, E-box proteins, CREB (cAMP response element binding protein), JUN/FOS, and E2F.
Equally important are inhibitors of Smad activity. Members of the Ski family of protooncogenes, SKI and SnoN, have been shown to antagonize TGF-β signaling via direct interaction with the Co-Smad, Smad4, and the R-Smads. Smads also bind to the transcriptional co-repressor TGIF. TGIF recruits histone deacetylase (HDAC) and other repressors to transcriptional complexes resulting in transcriptional repression.
Termination of Smad signaling occurs via dephosphorylation of the R-Smads as well as via ubiquitylation and proteasome-mediated degradation. As discussed above, Smurf proteins target Smads for ubiquitylation and thus, destruction. The complete characterization of which phosphatase dephosphorylates which phosphorylation site in a given R-Smad has yet to be completed. However, for both Smad2 and Smad3 the dephosphorylation of Ser residues in the MH2 domain (the tail phosphorylation sites) has been shown to be catalyzed by PPM1A which is a member of the metal-dependent Ser/Thr phosphatase (PPM) family. PPM1A is also known as PP2Cα. The Ser/Thr phosphatase identified as PP2A (encoded by the PPP2CB gene) has also been shown to dephosphorylate Smad3 but only under conditions of hypoxia. Tail dephosphorylation of Smad1 has been proposed to be catalyzed by another member of the PPM family, PDP1. PDP1 is a mitochondrial phosphatase best characterized as a pyruvate dehydrogenase (PDH) phosphatase. Dephosphorylation of linker phosphorylation sites in Smad proteins appears to be catalyzed by phosphatases of the aspartate-based phosphatase family which includes FCP1 [transcription factor IIF (TFIIF)-associating component of RNA polymerase II] and SCP1 (small CTD phosphatase 1). The FCP1 protein is encoded by the CTDP1 gene and the SCP1 protein is encoded by the CTDSP1 gene.
