Last Updated: October 31, 2025
Introduction to Proto-oncogenes
Most, if not all, cancer cells contain genetic damage that appears to be the responsible event leading to tumorigenesis. The genetic damage present in a parental tumorigenic cell is maintained (i.e. not correctable) such that it is a heritable trait of all cells of subsequent generations. Genetic damage found in cancer cells is of two types:
1. Dominant and the genes have been termed proto-oncogenes. The distinction between the terms proto-oncogene and oncogene relates to the activity of the protein product of the gene. A proto-oncogene is a gene whose protein product has the capacity to induce cellular transformation given it sustains some genetic insult. An oncogene is a gene that has sustained some genetic damage and, therefore, produces a protein capable of cellular transformation.
The process of activation of proto-oncogenes to oncogenes can include retroviral transduction or retroviral integration (see below), point mutations, insertion mutations, gene amplification, chromosomal translocation and/or protein-protein interactions.
Proto-oncogenes can be classified into many different groups based upon their normal function within cells or based upon sequence homology to other known proteins. As predicted, proto-oncogenes have been identified at all levels of the various signal transduction cascades that control cell growth, proliferation and differentiation. The list of proto-oncogenes identified to date is too lengthy to include here, therefore, only those genes that have been highly characterized are described. Proto-oncogenes that were originally identified as resident in transforming retroviruses were initially designated as c– indicative of the cellular origin as opposed to v– to signify original identification in retroviruses.
2. Recessive and the genes variously termed tumor suppressors, growth suppressors, recessive oncogenes or anti-oncogenes.
Given the complexity of inducing and regulating cellular growth, proliferation and differentiation, it was suspected for many years that genetic damage to genes encoding growth factors, growth factor receptors and/or the proteins of the various signal transduction cascades would lead to cellular transformation. This suspicion has proven true with the identification of numerous genes, whose products function in cellular signaling, that are involved in some way in the genesis of the tumorigenic state. The majority of these proto-oncogenes were identified by either of two means: as the transforming genes (oncogenes) of transforming retroviruses or through transfection of DNA from tumor cell lines into non-transformed cell lines and screening for resultant tumorigenesis.
Viruses and Cancer
Tumor cells also can arise by non-genetic means through the actions of specific tumor viruses. Tumor viruses are of two distinct types. There are viruses with DNA genomes (e.g. papilloma and adenoviruses) and those with RNA genomes (termed retroviruses).
RNA tumor viruses are common in chickens, mice and cats but rare in humans. The only currently known human retroviruses are the human T-cell leukemia viruses (HTLVs) and the related retrovirus, human immunodeficiency virus (HIV).
Retroviruses can induce the transformed state within the cells they infect by two mechanisms. Both of these mechanisms are related to the life cycle of these viruses. When a retrovirus infects a cell its RNA genome is converted into DNA by the viral encoded RNA-dependent DNA polymerase (reverse transcriptase). The DNA then integrates into the genome of the host cell where it can remain being copied as the host genome is duplicated during the process of cellular division. Contained within the sequences at the ends of the retroviral genome are powerful transcriptional promoter sequences termed long terminal repeats (LTRs). The LTRs promote the transcription of the viral DNA leading to the production of new virus particles.
At some frequency the integration process leads to rearrangement of the viral genome and the consequent incorporation of a portion of the host genome into the viral genome. This process is termed transduction. Occasionally this transduction process leads to the virus acquiring a gene from the host that is normally involved in cellular growth control. Because of the alteration of the host gene during the transduction process as well as the gene being transcribed at a higher rate due to its association with the retroviral LTRs the transduced gene confers a growth advantage to the infected cell. The end result of this process is unrestricted cellular proliferation leading to tumorigenesis. The transduced genes are termed oncogenes. The normal cellular gene in its unmodified, non-transduced form is termed a proto-oncogene since it has the capacity to transform cells if altered in some way or expressed in an uncontrolled manner. Numerous oncogenes have been discovered in the genomes of transforming retroviruses.
The second mechanism by which retroviruses can transform cells relates to the powerful transcription promoting effect of the LTRs. When a retrovirus genome integrates into a host genome it does so randomly. At some frequency this integration process leads to the placement of the LTRs close to a gene that encodes a growth regulating protein. If the protein is expressed at an abnormally elevated level it can result in cellular transformation. This is termed retroviral integration induced transformation. It has recently been shown that HIV induces certain forms of cancers in infected individuals by this integration induced transformation process.
Cellular transformation by DNA tumor viruses, in most cases, has been shown to be the result of protein-protein interaction. Proteins encoded by the DNA tumor viruses, termed tumor antigens or T antigens, can interact with cellular proteins. This interaction effectively sequesters the cellular proteins away from their normal functional locations within the cell. The predominant types of proteins that are sequestered by viral T antigens have been shown to be of the tumor suppressor type. It is the loss of their normal suppressor functions that results in cellular transformation.
Classifications of Proto-Oncogenes
Although there are numerous examples of each classification of proto-oncogene, the lists here are by no means exhaustive. New genes with tumor causing capabilities are being isolated continuously. Additional details on genes and their functions for the various categories listed below can be found in the Signal Transduction Pathways: Overview page.
Growth Factors
The beta polypeptide of the platelet-derived growth factor (PDGFB) was originally identified as the c-sis gene (the v-sis gene is the oncogene in simian sarcoma virus). The v-sis gene was the first viral oncogene to be identified as having homology to a known cellular gene.
The fibroblast growth factor 3 (FGF3) gene was originally identified as the int-2 gene, so named for the fact that it is a common site of integration of mouse mammary tumor virus.
The FGF4 gene was originally identified as an FGF-related growth factor in Kaposi’s sarcoma cells isolated from patients with AIDS and also identified in certain gastric carcinomas.
Receptor Tyrosine Kinases
Growth factor receptors are of several different types dependent upon whether or not they exhibit intrinsic enzymatic activity upon growth factor binding and what type of intrinsic activity is associated with the receptor. Numerous growth factor receptors exhibit intrinsic tyrosine kinase activity and many receptors in this class have been shown to have oncogenic potential.
The FMS (“fims”) gene encodes the colony stimulating factor-1 (CSF-1) receptor (CSF1R) and was first identified as a retroviral oncogene. The FLG (“flag”) gene, named because it has homology to the FMS gene, hence fms-like gene, encodes the fibroblast growth factor 1 receptor, FGFR1.
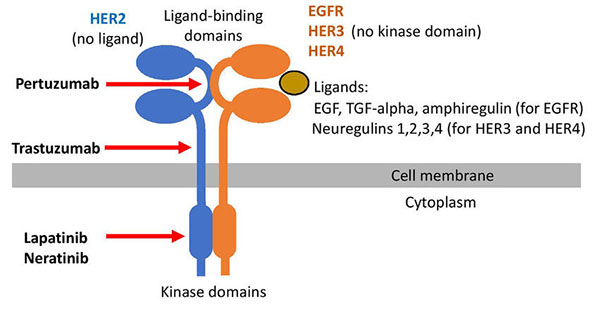
The epidermal growth factor receptor (EGFR, also called HER1 for human EGF receptor 1) is amplified in numerous cancers, in particular in squamous cell carcinomas. The NEU (“new”) gene was identified as an EGF receptor-related gene in an ethylnitrosourea-induced neuroblastoma. The NEU gene is officially identified as the ERB-B2 receptor tyrosine kinase 2 (ERBB2) and was also identified as HER2 (human EGF receptor 2). The conversion of proto-oncogenic to oncogenic NEU requires only a single amino acid change in the transmembrane domain.
The tropomyosin receptor kinase (TRK: “track”) genes encode the neurotrophin receptors (nerve growth factor receptor-like proteins). The first TRK gene was found in a pancreatic cancer. Subsequently, two additional TRK-related genes were identified. These three are now identified as TRKA, TRKB and TRKC. The only neurotrophin that binds and activates TRKA is simply neurotrophin or more commonly known as nerve growth factor, NGF. Several neurotrophins bind and activate TRKB including neurotrophin-3 (NT-3), NT-4, and brain-derived neurotrophic factor (BDNF). The only neurotrophin that activates TRKC is neurotrophin-3, NT-3.
The MET proto-oncogene encodes the hepatocyte growth factor (HGF)/scatter factor (SF) receptor.
The KIT gene, which encodes the stem cell factor receptor (SCFR) is found constitutively activated in sarcomas. SCF is also known as the mast cell growth factor. The viral KIT (v-kit) gene was originally identified in feline sarcomas.
Membrane Associated Non-Receptor Tyrosine Kinases
The SRC gene was the first identified oncogene. The SRC gene is the archetypal protein tyrosine kinase.
The LCK gene was isolated from a T cell tumor line (LYSTRA cell kinase) and has been shown to be associated with the CD4 and CD8 antigens of T cells.
The chromosome (chromosome 9) containing the ABL proto-oncogene (first identified in the Abelson murine leukemia virus) is frequently rearranged in chronic myelogenous leukemias (CMLs). The majority of rearrangements involve a reciprocal translocation between the ABL chromosome and chromosome 22 near a locus termed the break-point cluster region (BCR). The result is a constitutively active ABL tyrosine kinase domain fused to the BCR coding region forming what is referred to as the BCR-ABL fusion protein. This translocation is called the Philadelphia chromosome (Ph+). The anti-cancer drug imatinib (Gleevec®) targets the tyrosine kinase activity of the BCR-ABL protein in CML.
G-Protein Coupled Receptors
The MAS1 gene was identified in a mammary carcinoma and has been shown to be the receptor for the angiotensin-II metabolite identified as angiotensin-(1-7). When angiotensin-(1-7) binds and activates MAS1 the result is interference with the signaling from the angiotensin-II activated angiotensin receptor (AT1R).
Membrane Associated G-Proteins
There are numerous proteins in the cell whose function is regulated by the binding and hydrolyzing of GTP. These proteins all belong to a class of protein called G-proteins. In addition to the G-proteins themselves, their activity is regulated by associated GTPase-activating (GAP) proteins and guanine nucleotide exchange factors, (GEF).
Mutations in the RAS gene have been identified in a variety of different type of tumor cells. Indeed, the RAS gene is one of the most frequently disrupted genes in colorectal carcinomas as well as many other human cancers. The RAS gene represents the archetype of a large family of monomeric G-proteins that includes more than 100 family members. There are three primary members of the RAS family identified as HRAS, KRAS, and NRAS.
Genes of the G-protein regulator (GEF) family that have been identified as oncogenes are DBL and the three members of the VAV family, VAV1, VAV2, and VAV3. The official designation for the DBL gene is MCF.2 cell line-derived transforming sequence, MCF2. The MCF2-encoded protein is involved in the exchange of GTP for GDP in several proteins of the small G-protein class. These include RHO, RAC, and CDC42 (CDC = cell division cycle). The VAV proteins act as GEFs for members of the RHO family of monomeric G-proteins. The VAV1-encoded protein plays a role in the development and activation of T-cells and B-cells. The VAV2 gene is exclusively expressed in cells of the hematopoietic lineage. The VAV3-encoded protein in involved in actin cytoskeletal rearrangements and is also involved in alterations in transcriptional activity.
Serine/Threonine Kinases
The RAF gene family of encoded proteins are involved in the signaling pathways of most RTKs. There are three family members, RAF-1, A-RAF, and B-RAF. The RAF1-encoded protein is a MAP kinase kinase kinase (MAP3K) whose primary substrates are the dual-specificity kinases MEK1 and MEK2. The BRAF-encoded protein is involved in the MAP kinase/ERK signaling pathways.
The MOS gene (originally identified in the Moloney murine sarcoma virus) is normally expressed in germ cells and functions during oocyte maturation. The principal substrate for the MOS-encoded protein is the MAP kinase activator MEK1 (formally identified as MAP2K1).
Nuclear DNA-Binding/Transcription Factors
Many genes that are altered resulting in cancer encode proteins that are transcription factors. It would seem obvious that a genetic alteration that disrupts the normal expression and regulation of a transcription factor could have profound implications for cellular regulation.
The MYC gene was originally identified in the avian myelocytomatosis virus. A disrupted human MYC gene has been found to be involved in numerous hematopoietic neoplasias. Disruption of MYC has been shown to be the result of retroviral integration and transduction as well as chromosomal rearrangements. There are three MYC genes, each of which has been shown to be involved in cancer: MYC, N-MYC, and L-MYC.
The FOS gene was identified by Dr. Tom Curran as the transforming gene in the Finkel-Biskis-Jinkins (FBJ) murine osteogenic sarcoma virus. The protein interacts with a second proto-oncogenic protein, JUN to form a transcriptional regulatory complex.
The p53 gene was originally identified as a major nuclear antigen in transformed cells. The p53 gene is the single most identified mutant protein in human tumors. Mutant forms of the p53 protein interfere with cell growth suppressor effects of wild-type p53 indicating that the p53 gene product is actually a tumor suppressor.
Breast and Ovarian Cancer Susceptibility Genes
Not all breast and ovarian cancers are caused by inherited mutations in specific genes. However, several genetic loci have been shown to predispose an individual to these types of cancer. To date seven loci have been identified that when mutated predispose an individual to breast and ovarian cancer. These loci are BRCA1, BRCA2, ATM, CHEK2, BRIP1, PALB2, and RAD51C. Of significance is that all of these breast cancer susceptibility genes encode proteins that function in some aspect of DNA damage repair. In addition, several of these genes (BRCA2, PALB2, and BRIP1) are associated with the inheritance of Fanconi anemia (FA). FA is a rare childhood chromosomal instability disorder characterized by developmental abnormalities, bone marrow failure, and a predisposition to leukemias and other cancers. Several of these genes are included in the Hereditary Cancer Syndromes Table below.
ATM: This gene is called ataxia telangiectasia mutated. It is the gene first identified as causing ataxia telangiectasia.
BRCA1: This gene was originally called breast cancer 1, early onset. The designation now used is BRCA1 DNA repair associated. Along with BRCA2, these were the first genes identified as being mutated in inherited forms of breast and ovarian cancer. The BRCA1 gene is located on chromosome 17q21.31.
BRCA2: This gene was originally called breast cancer 2, early onset. The designation now used is BRCA2 DNA repair associated. This locus is also known as the Fanconi anemia complementation group D1 (FANCD1) gene. Of the three FA loci shown to have an association with breast cancer susceptibility only the BRCA2 gene plays a major role in high-risk breast cancer predisposition. The BRCA2 gene is located on chromosome 13q13.1.
BRIP1: This gene is called BRCA1 interacting protein C-terminal helicase 1. BRIP1 is a member of the RecQ DEAH family of RNA helicases. The protein functions in DNA double-strand break (DSB) repair in concert with BRCA1. The gene maps to chromosome 17q23.2. This locus is also known as the Fanconi anemia complementation group J (FANCJ) gene.
CHEK2: This gene is called cell cycle checkpoint kinase 2. CHEK2 is a serine threonine kinase that is activated by ATM protein in response to DNA double-strand breaks. CHEK2 not only regulates the function of BRCA1 protein in DNA repair but also exerts a number of critical roles in cell cycle control and apoptosis. The CHEK2 gene is located on chromosome 22q12.1.
PALB2: This gene is called partner and localizer of BRCA2 and encodes a protein that interacts with BRCA2 protein in the nucleus. PALB2 binds directly to BRCA1 and serves as the molecular scaffold in the formation of the BRCA1-PALB2-BRCA2 complex. The gene maps to chromosome 16p12.2. This locus is also known as the Fanconi anemia complementation group N (FANCN) gene.
RAD51C: This gene is a member of the RAD51 family of related genes that encode proteins involved in recombinational DNA repair and meiotic recombination. The RAD51C gene is essential for homologous DNA recombination repair. The gene is located on chromosome 17q22.
Table of Hereditary Cancer Syndromes
| Syndrome | Cloned Gene | Function | Chromosomal Location | Tumor Types |
| Li-Fraumeni Syndrome | TP53 | tumor suppressor; cell cycle regulation, apoptosis | 17p13.1 | brain tumors, sarcomas, leukemia, breast cancer |
| Familial Retinoblastoma | RB1 | tumor suppressor; cell cycle regulation | 13q14.2 | retinoblastoma, osteogenic sarcoma |
| Wilms Tumor | WT1 | tumor suppressor; transcriptional regulation | 11p13 | pediatric kidney cancer |
| Neurofibromatosis Type 1 | NF1; protein is neurofibromin 1 | tumor suppressor; catalysis of RAS inactivation | 17q11.2 | neurofibromas, sarcomas, gliomas |
| Neurofibromatosis Type 2 | NF2; protein is merlin, also called neurofibromin 2 | tumor suppressor; linkage of cell membrane to cytoskeleton | 22q12.2 | Schwann cell tumors, astrocytomas, meningiomas, ependynomas |
| Familial Adenomatous Polyposis | APC | tumor suppressor; signaling through adhesion molecules to nucleus | 5q22.2 | colon cancer |
| Tuberous sclerosis 1 | TSC1= tumor suppressor protein = hamartin | tumor suppressor; forms complex with TSC2 protein, inhibits signaling to downstream effectors of mTOR | 9q34 | seizures, intellectual impairment, facial angiofibromas |
| Tuberous sclerosis 2 | TSC2; protein is tuberin | tumor suppressor; see TSC1 above | 16p13.3 | benign growths (hamartomas) in many tissues, astrocytomas, rhabdomyosarcomas |
| Deleted in Pancreatic Carcinoma 4 | DPC4; also known as SMAD4 | tumor suppressor; regulation of TGF-β/BMP signal transduction | 18q21.1 | pancreatic carcinoma, colon cancer |
| Deleted in Colorectal Carcinoma | DCC | tumor suppressor; transmembrane receptor involved in axonal guidance via netrins | 18q21.2 | colorectal cancer |
| Familial Breast Cancer | BRCA1 | tumor suppressor; functions in transcription, DNA binding, transcription coupled DNA repair, homologous recombination, chromosomal stability, ubiquitylation of proteins, and centrosome replication | 17q21.31 | breast and ovarian cancer |
| Familial Breast Cancer | BRCA2; same as the FANCD1 locus (see below) | tumor suppressor; transcriptional regulation of genes involved in DNA repair and homologous recombination | 13q12.3 | breast and ovarian cancer |
| Peutz-Jeghers Syndrome (PJS) | STK11; protein is serine-threonine kinase 11, was also known as LKB1 | tumor suppressor; phosphorylates and activates AMP-activated kinase (AMPK), AMPK involved in stress responses, lipid and glucose metabolism | 19p13.3 | hyperpigmentation, multiple hamartomatous polyps, colorectal, breast and ovarian cancers |
| Lynch Syndrome; also known as Hereditary Nonpolyposis Colorectal Cancer (HNPCC) | MLH1: mutL homolog 1 MSH2: mutS homolog 2 MSH6: mutS homolog 6 PMS2: postmeiotic segregation increased 2 EPCAM: epithelial cell adhesion molecule | tumor suppressors; DNA mismatch repair; mutations at the end of EPCAM gene disrupt the function of the closely associated MSH2 gene | MLH1: 3p22.2 MSH2: 2p21–p16.3 MSH6: 2p16.3 PMS2: 7p22.1 EPCAM: 2p21 | colorectal, endometrial, ovarian, stomach, small intestine, pancreatic, and urinary tract cancers |
| von Hippel-Lindau Syndrome | VHL | tumor suppressor; is a component of a ubiquitin ligase complex that ubiquitylates HIF-1α; also involved in inhibition of transcription elongation | 3p25.3 | renal cancers, hemangioblastomas, pheochromocytoma |
| Familial Melanoma | CDKN2A; protein is cyclin-dependent kinase inhibitor 2A; gene produces 2 proteins: p16INK4 and p14ARF | tumor suppressor; p16INK4 inhibits cell-cycle kinases CDK4 and CDK6; p14ARF binds the p53 stabilizing protein MDM2 | 9p21 | melanoma, pancreatic cancer, others |
| Gorlin Syndrome: Nevoid basal cell carcinoma syndrome (NBCCS) | PTCH; protein is patched | tumor suppressor; transmembrane receptor for sonic hedgehog (SHH), involved in early development through repression of action of the frizzled class of GPCR called smoothened (SMO) | 9q22.3 | basal cell skin cancer |
| Multiple Endocrine Neoplasia Type 1 | MEN1; protein is menin | tumor suppressor; serves as a scaffold protein for processes of histone modification | 11q13.1 | parathyroid and pituitary adenomas, islet cell tumors, carcinoid |
| Multiple Endocrine Neoplasia Type 2 | MEN2; also known as RET (meaning rearranged during transfection) | transmembrane receptor tyrosine kinase for glial-derived neurotrophic factor (GDNF) | 10q11.2 | medullary thyroid cancer, type 2A pheochromocytoma, mucosal hamartoma |
| Beckwith-Wiedemann Syndrome (BWS) | BWS caused by changes in a 1 megabase region that encompasses at least 15 genes: CDKN1C (also called p57KIP2) is likely responsible for the cancers | CDKN1C is a cyclin-dependent kinase inhibitor, cell cycle regulator | 11p15.5 | genomic imprinting disorder resulting in Wilms tumor, adrenocortical cancer, hepatoblastoma |
| Hereditary papillary renal cancer (HPRC) | MET | transmembrane receptor for hepatocyte growth factor (HGF) | 7q31 | renal papillary cancer |
| Cowden syndrome | PTEN = phosphatase and tensin homolog | tumor suppressor; contains a catalytic domain similar to tyrosine phosphatases but is specific for phosphatidylinositides with the primary activity being phosphatidylinositol-3,4,5-trisphophate (PIP3) 3-phosphatase; PTEN activity serves to negatively regulate the AKT/PKB signaling pathway | 10q23.3 | breast cancer, thyroid cancer, head and neck squamous carcinomas |
| Hereditary prostate cancer, numerous loci: HPC1(PRCA1), HPCX, MXI1, KAI1, PCAP | HPC1 and PRCA1 are same designation, ribonuclease L (RNaseL) maps to this locus | RNaseL involved in mRNA degradation | 1q24-q25 | prostate cancer |
| Ataxia telangiectasia (AT) | ATM: 4 complementation groups: ATA, ATC, ATD, ATE, are associated with mutations in the ATM gene | gene product encodes a kinase, one substrate is p53 | 11q22.3 | lymphoma, cerebellar ataxia, immunodeficiency |
| Bloom syndrome | BLM | DNA helicase RecQ protein-like-3 | 15q26.1 | solid tumors, immunodeficiency |
| Xeroderma pigmentosum (XP), 8 complentation groups | XPA, ERCC3 (XPB), XPC, ERCC2 (XPD), DDB2 (XPE), ERCC4 (XPF), ERCC5 (XPGC), and variant XP (XPV, POLH gene) | DNA repair helicases; nucleotide excision repair; the ERCC genes are excision repair cross-complementation group genes; DDB2 is damage specific DNA binding protein 2; POLH is DNA polymerase eta (η) | A: 9q22.3 B: 2q21 C: 3p25.1 D: 19q13.3 E: 11p11.2 F: 16p13.3 GC: 13q22–q34 POLH: 6p21 | skin cancer |
| Fanconi anemia: 21 genes within the complementation groups; 10 are common | FANCA, B, C, BRCA2 (D1), D2, E, F, G, I, BRIP1 (J), L, M, PALB2 (N), RAD51C (O), SLX4 (P), ERCC4 (Q), RAD51 (R), BRCA1 (S), UBE2T (T), XRCC2 (U), MAD2L2 (V), RFWD3 (W) FANCA, C, E, F, G, and L form nuclear multiprotein complex | components of DNA repair machinery; BRIP1: BRCA1 interacting protein C-terminal helicase 1; PALB2: partner and localizer of BRCA2; RAD51C: RAD51 paralog C; SLX4: SLX4 structure-specific endonuclease subunit; ERCC4: excision repair cross-complementation group 4; UBE2T: ubiquitin conjugating enzyme E2T; XRCC2: X-ray repair cross complementing 2; MAD2L2: mitotic arrest deficient 2 like 2; RFWD3: RING finger and WD repeat domain 3 FA-M, FA-O, FA-T, FA-U, FA-V, FA-W variants only found in a single individual; FA-Q, FA-R variants only found in two individuals | 10 common alleles only A: 16q24.3 B: Xp22.2 C: 9q22.3 D1: 13q13.1 D2: 3p25.3 E: 11p15 F: 11p14.3 G: 9p13 I: 2p16.1 J: 17q23.2 | Cancers: acute myeloid leukemia (AML), adult onset aplastic anemia, myelodysplastic syndrome (MDS) Other features: chromosomal instability, thrombocytopenia, leukopenia, neutropenia, abnormal skin pigmentation, ophthalmic anomalies, skeletal malformations |