Last Updated: December 16, 2025
Sources of NADPH
The reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) is critical to numerous reductive biosynthetic processes such as fatty acid synthesis, cholesterol synthesis, bile acid synthesis, steroid hormone synthesis, and deoxynucleotide synthesis. NADPH also is used in the processes of cellular protection against redox stress by providing reducing equivalents to antioxidants such as glutathione and thioredoxin. The significance of NADPH levels to cellular survival have been demonstrated by overexpression of several cytoplasmic enzymes that synthesize NADPH leading to lifespan extension in model organisms such as Drosophila melanogaster. Mitochondrial NADPH plays an important role in the protection against redox stress and cell death and the levels of at least one mitochondrial NADPH-utilizing enzyme, thioredoxin reductase 2, correlate with cellular longevity.
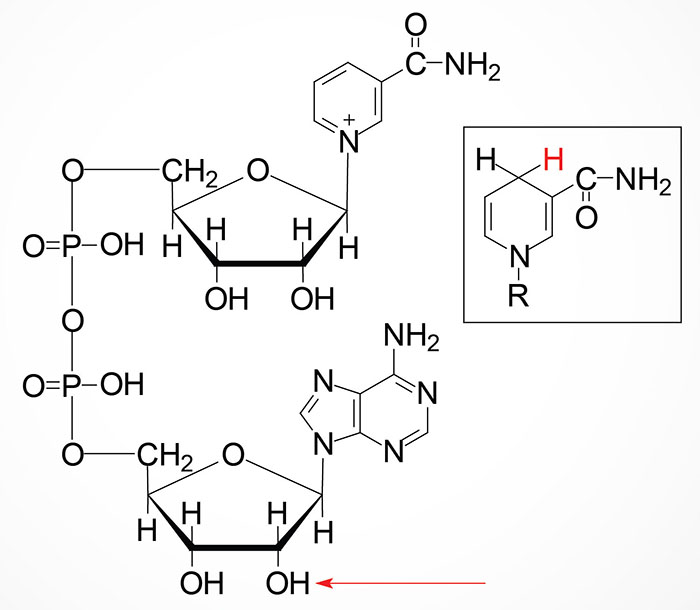
The details of the synthesis of nicotinamide adenine dinucleotide (NAD+) can be found in the Vitamin B3: Metabolism and Functions page. NAD+ can be converted to NADP+ through the action of cytoplasmic NAD+ kinase (NADK) or mitochondrial NAD+ kinase 2 (NADK2). The NADK gene is located on chromosome 1p36.33 and is composed of 19 exons that generate four alternatively spliced mRNAs that collectively generate three distinct isoforms of the enzyme. The NADK2 gene is located on chromosome 5p13.2 and is composed of 15 exons that generate four alternatively spliced mRNAs, that collectively encode three distinct isoforms of the enzyme.
Given the significant roles of NADPH in numerous pathways of reductive biosynthesis and in the maintenance of cellular redox status, in both the cytosol and the mitochondria, it is not surprising that there are multiple cytoplasmic and mitochondrial enzymes that have, as a product of their reactions, the reduced electron carrier, NADPH.
Cytoplasmic NADPH Generating Pathways
Pentose Phosphate Pathway Enzymes
Several important biochemical pathways, that generate NADPH, are not directly related to NAD+ synthesis and salvage. As indicated above, NADP+ can be generated via the action of cytoplasmic and mitochondrial NAD+ kinases. The reduction of NADP to NADPH occurs in the course of various oxidation reactions. The Pentose Phosphate Pathway (PPP) is one of the most significant metabolic pathways for NADPH generation. Two of the enzymes of the oxidative portion of the PPP generate NADPH in the course of oxidizing glucose-6-phosphate. These enzymes are glucose-6-phosphate dehydrogenase (encoded by the G6PD gene) and 6-phosphogluconate dehydrogenase (encoded by the PGD gene).
Malic Enzymes (ME)
The synthesis of NADPH via the action of malic enzyme is directly coupled to the process of the transport of acetyl-CoA from the mitochondria to the cytosol where the acetyl-CoA can then be used for fatty acid synthesis, cholesterol synthesis, phospholipid synthesis, and deoxynucleotide synthesis as indicated above.
Humans express three malic enzymes, one cytoplasmic that requires NADP+ and two mitochondrial enzymes, one that requires NADP+ and one that requires NAD+. The NADP+-dependent cytoplasmic enzyme is called malic enzyme 1 and is encoded by the ME1 gene. The ME1 gene is located on chromosome 6q14.2 and is composed of 14 exons that encode a protein of 572 amino acids.
Isocitrate Dehydrogenases (IDH)
Humans express three different IDH enzymes identified as IDH1, IDH2, and IDH3. The IDH3 isoform is involved in the TCA cycle where it oxidatively decarboxylates isocitrate to 2-oxoglutarate (α-ketoglutarate). Both IDH1 and IDH2 enzymes utilize NADP+ as their cofactor and generate NADPH via oxidative decarboxylation of isocitrate outside the context of the TCA cycle. The IDH1 enzyme is localized to the cytosol and the peroxisomes, whereas IDH2 is localized to the mitochondria.
The IDH1 is a homodimeric enzyme encoded by the IDH1 gene located on chromosome 2q33.3 and is composed of 12 exons that generate three alternatively spliced mRNAs, all of which encode the same 414 amino acid protein. IDH1 is found primarily in the cytosol and the peroxisomes.
The primary functions of IDH1 and IDH2 are to serve as producers of NADPH to enhance the cellular responses to oxidative stress and the generation of reactive oxygen species, ROS. The significance of IDH1 and IDH2 produced NADPH can be made clear by pointing out that cells with low level IDH1 and IDH2 expression are more sensitive to oxidative stress than cells with higher levels of expression of these two enzymes.
Cytoplasmic and Mitochondrial Tetrahydrofolate Interconversions
A member of the folate metabolism family of enzymes, encoded by the MTHFD1 gene, can generate NADPH in the course of the conversion of N5,N10-methylene-tetrahydrofolate (5,10-methylene-THF) to 5,10-methenyl-THF. The MTHFD1 encoded enzyme possesses three activities that include, N5,N10-methenyl-THF (5,10-methenyl-THF) cyclohydrolase, 5,10-methylene-THF dehydrogenase, and 10-formyl-THF synthetase.
Two additional enzymes of folate metabolism, aldehyde dehydrogenase 1 family member L1 (encoded by the ALDH1L1 gene) and ALDH1L2, produce NADPH in the course of converting N10-formyltetrahydrofolate (10-formyl-THF) to tetrahydrofolate (THF) and CO2. The ALDH1L1 encoded enzyme is localized to the cytosol and the ALDH1L2 encoded enzyme is localized to the mitochondria. The ALDH1L1 encoded enzyme was originally identified as 10-formyl tetrahydrofolate dehydrogenase (FTHFD; also designated 10-FTHFDH).
Endoplasmic Reticulum (ER) NADPH Generating Pathways
Hexose-6-Phosphate Dehydrogenase (H6PD)
In addition to the cytosolic, pentose phosphate pathway-associated glucose-6-phosphate dehydrogenase activity, humans express an endoplasmic reticulum (ER) and the sarcoplasmic reticulum (SR) localized glucose-6-phosphate dehydrogenase activity referred to as the H form. This form of glucose-6-phosphate dehydrogenase activity is identified as hexose-6-phosphate dehydrogenase (encoded by the H6PD gene) and also as glucose 1-dehydrogenase. Within the ER, hexose-6-phosphate dehydrogenase converts glucose-6-phosphate and NADP+ to 6-phosphogluconate and NADPH. In addition to glucose-6-phosphate, H6PD can metabolize other hexose-6-phosphates, glucose-6-sulfate, and glucose.
One of the primary functions of the ER- and SR-localized NADPH is to maintain redox homeostasis within these organelles. Loss of ER redox homeostasis can lead to ER stress and induction of the unfolded protein response (UPR) which, if severe enough will trigger cell death via the apoptotic pathway.
Another principal function of the NADPH produced by ER-localized hexose-6-phosphate dehydrogenase is to provide the reducing energy to ER-localized reductases, specifically those involved in steroid hormone metabolism, with 11β-hydroxysteroid dehydrogenase 1 (11β-HSD1; encoded by the HSD11B1 gene) being particularly important. The primary function of the HSD11B1 encoded enzyme is to reduce the 11-oxo groups in cortisone and 11-dehydrocorticosterone to the active glucocorticoids, cortisol and corticosterone, respectively. However, the enzyme can, under certain conditions, also inactivate cortisol and corticosterone by catalyzing the oxidation reactions converting cortisol to cortisone and corticosterone to 11-dehydrocorticosterone. Of clinical significance to the role of ER-localized NADPH is that mutations in the H6PD gene are associated with glucocorticoid deficiency.
Mitochondrial NADPH Generating Pathways
Malic Enzymes (ME)
The NADP+-dependent mitochondrial malic enzyme is called malic enzyme 3 and is encoded by the ME3 gene. The ME3 gene is located on chromosome 11q14.2 and is composed of 22 exons that generate four alternatively spliced mRNAs that all encode the same 604 amino acid protein.
Isocitrate Dehydrogenases (IDH)
The IDH2 enzyme is, like IDH1, a homodimeric enzyme but it is located to the mitochondria. IDH2 is encoded by the IDH2 gene located on chromosome 15q26.1 and is composed of 12 exons that generate three alternatively spliced mRNAs, each of which encode a distinct protein isoform.
Mitochondrial Tetrahydrofolate Interconversions
One of the enzymes of folate metabolism, aldehyde dehydrogenase 1 family member L2, produces NADPH in the course of converting N10-formyltetrahydrofolate (10-formyl-THF) to tetrahydrofolate (THF) and CO2. This aldehyde dehydrogenase family member is localized to the mitochondria where it performs the same reaction as the cytosolic version encoded by the ALDH1L1 gene. Aldehyde dehydrogenase 1 family member L2 (encoded by the ALDH1L2 gene) was originally identified as mitochondrial 10-formyltetrahydrofolate dehydrogenase.
Another mitochondria localized member of the folate metabolizing enzymes is methylene tetrahydrofolate dehydrogenase (NADP+ dependent) 1-like which is encoded by the MTHFD1L gene. Unlike the MTHFD1 encoded enzyme that is a tri-functional enzyme harboring N5,N10-methenyl-THF (5,10-methenyl-THF) cyclohydrolase, 5,10-methylene-THF dehydrogenase, and 10-formyl-THF synthetase activities, the MTHFD1L encoded enzyme only possesses 10-formy-lTHF synthetase activity.
Another mitochondria localized member of the folate metabolizing enzymes is methylene tetrahydrofolate dehydrogenase (NADP+ dependent) 2-like which is encoded by the MTHFD2L gene. The MTHFD2L encoded enzyme is a bi-functional enzyme harboring N5,N10-methenyl-THF (5,10-methenyl-THF) cyclohydrolase and 5,10-methylene-THF dehydrogenase activities. The human MTHFD2 gene encoded enzyme is also a mitochondrial enzyme that harbors the same activities as MTHFD2L encoded enzyme. However, expression of the MTHFD2 gene is only found in transformed cells and in embryonic non-differentiated tissues.
Nicotinamide Nucleotide Transhydrogenase
Nicotinamide nucleotide transhydrogenase, which is encoded by the NNT gene, is embedded in the inner mitochondrial membrane. The NNT encoded enzyme utilizes the energy of the mitochondrial proton gradient (the proton motive force) to catalyze hydride transfer from NADH to NADP+ generating NADPH.
Roles of NADPH in Reductive Biosynthesis
NADPH in Fatty Acid Synthesis
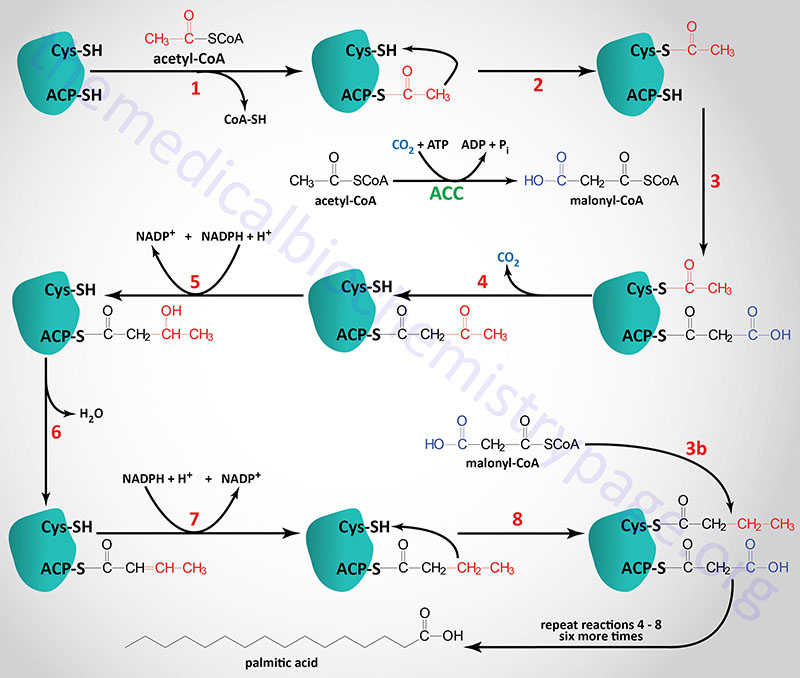
During the de novo synthesis of palmitic acid, fatty acid synthase (FAS) utilizes NADPH for two of the reactions of each cycle of acetyl-CoA addition. These reactions are the β-ketoacyl-ACP reductase and enoyl-CoA reductase catalyzed steps indicated as reaction 5 and 7 in the Figure below (details in the Synthesis of Fatty Acids page). Each mole of palmitic acid synthesized by FAS requires 12 moles of NADPH.
NADPH in Cholesterol Synthesis
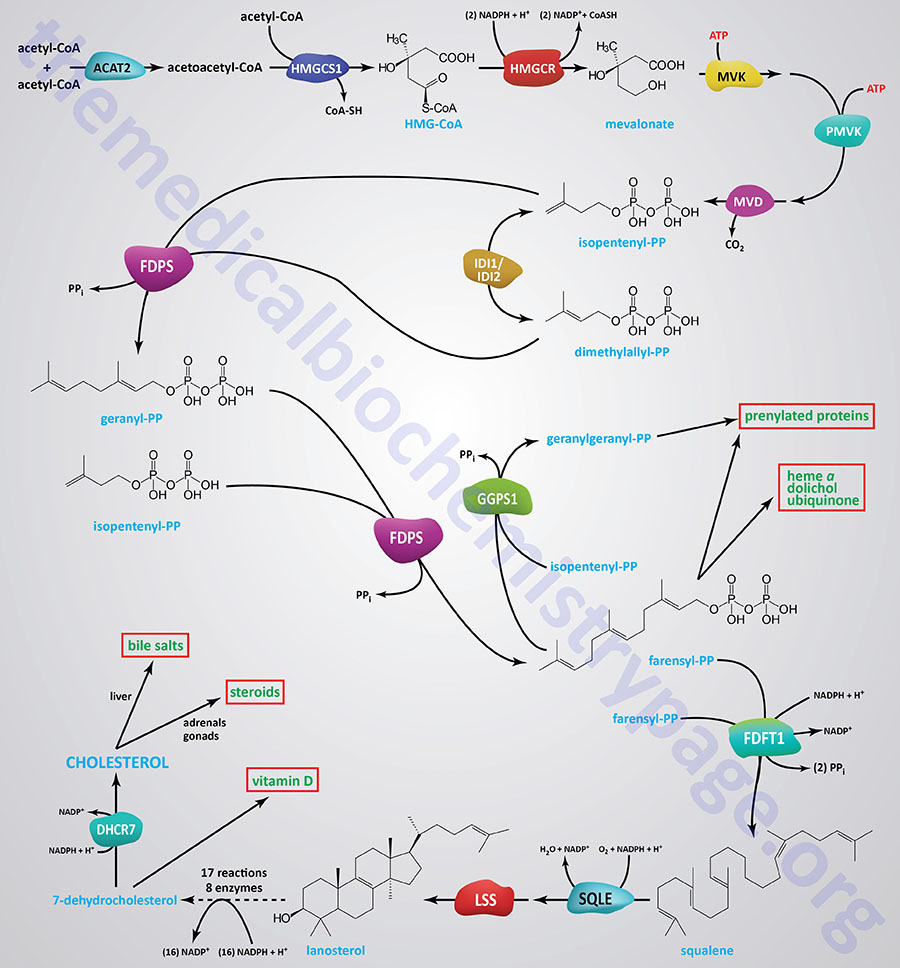
Several enzymes in the pathway of cholesterol synthesis require NADPH for their activities. The pathway of cholesterol synthesis is shown in the Figure below, however, several of the activities requiring NADPH are in the 18 reactions from lanosterol to 7-dehydrocholesterol that are not included in the Figure. The initial reactions of cholesterol synthesis, to the formation of isopentenyl pyrophosphate, are commonly referred to as the mevalonate pathway.
The most significant reaction of mevalonate pathway, which represents the rate-limiting reaction, is catalyzed by 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR; encoded by the HMGCR gene). The HMGR catalyzed reaction requires two mole of NADPH.
During the condensation of farnesylpyrophosphates to squalene, the enzyme farnesyl-diphosphate farnesyltransferase 1 (commonly called squalene synthase) utilizes a mole of NADPH during the reduction of a carbon-carbon double bond (-C=C-). During the epoxidation of squalene, catalyzed by squalene epoxidase (encoded by the SQLE gene; also called squalene monooxygenase), another mole of NADPH is utilized.
In the 16 reactions (catalyzed by 8 different enzymes) that constitute the conversion of lanosterol to 7-dehydrocholesterol there many that require NADPH such that a total of 15 moles of NADPH are consumed. Details of the reactions and enzymes of lanosterol conversion to 7-dehydrocholesterol are presented in the Cholesterol: Synthesis, Metabolism, and Regulation page.
The terminal reaction of cholesterol synthesis, the conversion of 7-dehydrocholesterol to cholesterol, is catalyzed by the NADPH -requiring enzyme, 7-dehydrocholesterol reductase (encoded by the DHCR7 gene).
NADPH in Steroidogenesis
The many pathways of steroid hormone synthesis, adrenal cortical and gonadal, that utilize NADPH are those involving the 11β-hydroxysteroid dehydrogenases (HSD11B) and the 17β-hydroxysteroid dehydrogenases (HSD17B). These enzymes can function in either direction, NADPH oxidation, or NADP+ reduction dependent upon the redox state.
A major enzyme of steroid hormone metabolism that utilizes NADPH is the ER-localized 11β-hydroxysteroid dehydrogenase 1 (11β-HSD1) which is encoded by the HSD11B1 gene. The primary function of the HSD11B1 encoded enzyme is to reduce the 11-oxo groups in cortisone and 11-dehydrocorticosterone to the active glucocorticoids, cortisol and corticosterone, respectively. However, the enzyme can, under certain conditions, also inactivate cortisol and corticosterone by catalyzing the oxidation reactions converting cortisol to cortisone and corticosterone to 11-dehydrocorticosterone. Of clinical significance to the role of ER-localized NADPH is that mutations in the H6PD gene are associated with glucocorticoid deficiency.
The HSD17B enzymes, all of which are members of the short chain dehydrogenase/reductase superfamily (SDR), includes 15 members, six of which utilize NADPH in the normal direction (reduction) of synthesis. The HSD17B1, HSD17B3, ARK1C3 (also known as HSD17B5), HSD17B7, HSD17B12, and RDH11 (also known as HSD17B15) enzymes all function in some capacity in the metabolism of steroids. The HSD17B1 and HSD17B3 encoded enzymes are involved in the synthesis of the male and female sex hormones.
NADPH in Control of Redox Status and Aging
NADPH-Coupled Redox Systems and Aging
The cellular levels of NADPH decrease with the aging process due to two primary forces. The level of NAD+ decreases with aging and the cumulative effects of oxidative stress ratios of NADP+ and NADPH gradually shifts to higher NADP+ relative to NADPH.
As we age the cellular NAD+ levels decline in part due to the reduced expression of one of the enzymes of NAD+ salvage, nicotinamide phosphoribosyl transferase (NAMPT) as well as due to increased NAD+ degradation via increased activity of the NAD+ and NADP+ hydrolyzing enzyme, CD38. Studies have shown that CD38 has a much higher affinity for NADP+ than for NAD+. Decreased NAD+ levels can contribute to sirtuin mediated deacetylase-dependent alterations in both mitochondrial electron transport activity (resulting in increased production of reactive oxygen species, ROS) as well as to increased nuclear DNA damage leading to the activation poly-ADP-ribose polymerases (PARP). Increased CD38 and PARP activity results in increased hydrolysis of NAD+ and NADP+. Activation of PARP-1 also leads to decreased NADPH levels because this particular PARP inhibits the glycolytic enzyme, hexokinase. Phosphorylation of glucose by hexokinase is not only required to drive glucose into the glycolytic pathway but also into the pentose phosphate pathway, the major NADPH generating metabolic pathway.
Decreased mitochondrial electron transport function with aging results in a decreased proton gradient across the inner mitochondrial membrane, the proton-motive force. As indicated above, the proton motive force is utilized by mitochondrial
nicotinamide nucleotide transhydrogenase (NNT) to catalyze hydride transfer from NADH to NADP+ generating NADPH.
As the [NADP+]/[NADPH] ratio, in both the cytosol and the mitochondria, increases with aging the activities of glutathione-disulfide reductase (encoded by the GSR gene; commonly called glutathione reductase) and thioredoxin reductases are reduced. Humans express both cytoplasmic thioredoxin reductase (encoded by the TXNRD1 gene) and mitochondrial (encoded by the TXNRD2 gene). The reduced levels of these enzymes leads to higher levels of the oxidized states of glutathione (written GSSG) and the thioredoxins reducing their roles in detoxification of reactive oxygen species (ROS) with the outcome being increased cellular oxidative damage.
NADPH in Erythrocyte Oxidative Control
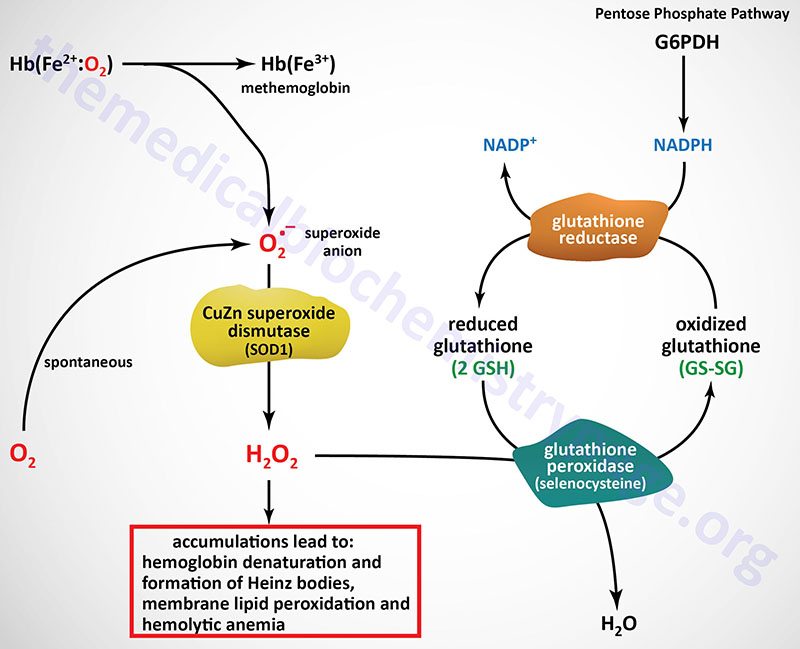
The predominant pathways of carbohydrate metabolism in the red blood cell (RBC) are glycolysis, the pentose phosphate pathway (PPP) and 2,3-bisphosphoglycerate (2,3-BPG) metabolism (refer to the Hemoglobin and Myoglobin page for review of the synthesis and role role of 2,3-BPG). Glycolysis provides ATP for membrane ion pumps and NADH for re-oxidation of methemoglobin. The PPP supplies the RBC with NADPH to maintain the reduced state of glutathione.
The inability to maintain reduced glutathione in RBCs leads to increased accumulation of peroxides, predominantly H2O2, that in turn results in a weakening of the cell wall as a result of membrane lipid peroxidation resulting in concomitant hemolysis. Accumulation of H2O2 also leads to increased rates of cysteine sulfhydryl oxidation in hemoglobin resulting in the formation of cross-linked complexes of denatured hemoglobin. These large complexes of denatured hemoglobin can be visualized by light microscopy and are referred to as Heinz bodies.
Glutathione removes peroxides from membrane lipids and serves as a substrate for H2O2 reduction to H2O via the action of glutathione peroxidase. The PPP in erythrocytes is the only pathway for these cells to produce NADPH, therefore, any defect in the production of NADPH, such as due to deficiencies in glucose-6-phosphate dehydrogenase, will have profound effects on erythrocyte survival.
The glutathione reductase gene (symbol: GSR) is located on chromosome 8p21.1 and is composed of 13 exons that generate four alternatively spliced mRNAs that encode four distinct protein isoforms. Humans express eight distinct glutathione peroxidase genes identified as GPX1–GPX8.
The GPX1 gene is located on chromosome 3p21.3 and is composed of 2 exons that generate two alternatively spliced mRNAs encoding two distinct protein isoforms.
The GPX2 gene is located on chromosome 14q24.1 and is composed of 4 exons that encode a 190 amino acid protein.
The GPX3 gene is located on chromosome 5q33.1 and is composed of 5 exons that encode a 226 amino acid precursor protein.
The GPX4 gene is located on chromosome 19p13.3 and is composed of 8 exons that generate three alternatively spliced mRNAs that encode several distinct protein isoforms that exhibit distinct subcellular sites of localization including the nucleus, the mitochondria, and the cytosol. In addition, one of the GPX4 encoded proteins plays a role in sperm maturation, a process that is structural not enzymatic.
The GPX5 gene is located on chromosome 6p22.1 and is composed of 7 exons that generate two alternatively spliced mRNAs encoding two protein isoforms. The GPX5 encoded mRNAs do not contain a selenocysteine (UGA) codon and, therefore, they encode selenium-independent enzymes. Expression of the GPX5 gene is restricted to the epididymis in the male gonads and the encoded proteins function to protect the membranes of spermatozoa during their maturation.
The GPX6 gene is located on chromosome 6p22.1 near the GPX5 gene and is composed of 5 exons that encode a 221 amino acid precursor protein. Expression of the GPX6 gene is restricted to the olfactory epithelium.
The GPX7 gene is located on chromosome 1p32 and is composed of 3 exons that encode a 187 amino acid precursor protein.
The GPX8 gene is located on chromosome 5q11.2 and is composed of 3 exons that encode a putative GPx enzyme.
NADPH in Detoxification
The liver is the major organ for the detoxification of endogenous and exogenous (xenobiotic) compounds. Many of the enzymes of hepatic xenobiotic metabolism are members of the cytochrome P450 family of enzymes that function as monooxygenases. The mitochondrial family of CYP enzymes utilize ferredoxin 1 (encoded by the FDX1 gene: protein also called adrenodoxin) and ferredoxin reductase (encoded by the FDXR gene; protein also called adrenodoxin reductase) in the transfer of electrons from NADPH to the cytochrome P450.