Last Updated: October 31, 2025
Introduction to Eukaryotic Cell Cycles
In 1858 the pathologist Rudolph Virchow coined the cell doctrine which states that “When a cell arises, there must have been a previous cell, just as animals can only arise from animals and plants from plants.” This doctrine is founded on the understanding that whether one is examining a single-celled organism or an animal as complex as man, the product is a result of repeated rounds of cell growth and division. Most eukaryotic cells will proceed through an ordered series of events (described in the section below) in which the cell duplicates its contents and then divides into two cells. This cycle of duplication and division is called the cell cycle.
In the context of somatic cell duplication and division where two identical daughter cells are produced both of which contain the same diploid content of DNA as the parental cell the process is referred to as mitosis. When diploid germ cells divide to produce haploid gametes (sex cells) the process is referred to as meiosis. In order to maintain the fidelity of the developing organism this process of cell division in multicellular organisms must be highly ordered and tightly regulated. The loss of control (as discussed in the sections below) will lead to abnormal development and is the cause of cancer.
The eukaryotic cell cycle is composed of a series of ordered steps, or phases, as depicted in the Figure below.
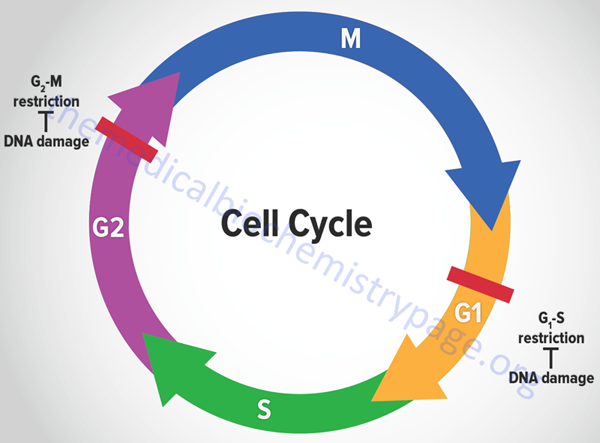
Of the four phases depicted in the Figure, the two critical steps are DNA replication, which occurs during S-phase, and the physical process of cell division which occurs during M-phase (for mitosis). If we start at the beginning of the process, a cell undergoes a period where all of the necessary machinery for the process of DNA replication is synthesized. This process occurs during what is referred to as a gap between S-phase and M-phase and is termed G1. Following DNA replication, the cell pauses in another gap phase termed G2 where all the machinery necessary for cell division is synthesized. M-phase is composed of two discreet steps: mitosis, which constitutes the pairing and separation of the duplicated chromosomes, and cytokinesis which is the physical process whereby the cell splits into two daughter cells. Not all cells continue to divide during the life-span of an organism. Many cells undergo what is referred to as terminal differentiation and become quiescent and no longer divide. Cells in this phase of their life-cycle are said to reside in another gap phase called G0. Under certain conditions, such as that resulting from an external signal stimulating cell growth, cells can exit the quiescent state and re-enter the cell cycle.
The Ordered Steps of Mitotic Cell Cycles
G1
The first gap in the normal cell cycle is called G1 and is the period when the necessary proteins for DNA replication are synthesized. However, this phase of the cell cycle is not only characterized by synthesis of replication machinery. During this period the cell must monitor both the internal and external environments to ensure that all the preparations for DNA synthesis have been completed and that overall conditions for cell division are favorable. As discussed below, there is a major check-point in a normal cell cycle that is critical for ensuring that all is well for the cell to enter S-phase.
S-Phase
The duplication of the cellular content of DNA occurs during S-phase, so-called because this is the phase when DNA is synthesized. This phase of the cell cycle is the longest taking 10–12 hours of a typical 24hr eukaryotic cell cycle. The amount of DNA in both mitotic and meiotic cells is defined by either “c” or “C” (for content) or “n” or “N” (for the number of complete sets of chromosome). In gametes (sperm and egg, which are haploid), the amount of DNA is 1C and the chromosome number is 1N. When egg and sperm fuse during fertilization the amount of DNA doubles to 2C and the chromosome number also doubles to 2N forming a diploid zygote. In mitotic cells, undergoing division to two identical daughter cells, the amount of DNA following S-phase is 4C but each pair of sister chromatids is still counted as a single chromosome such that the chromosome number remains 2N. In meiotic cells, following meiosis I, the gametes have a DNA content of 4C and a chromosome number of 2N. Following meiosis II the resulting four haploid gametes have a content of DNA of 1C and a chromosome number of 1N.
G2
During the second gap phase of the cell cycle the cell undertakes the synthesis of the proteins required to assemble the machinery required for separation of the duplicated chromosomes (the process called mitosis) and ultimately division of the parental cell into two daughter cells (the process termed cytokinesis). Like the G1 phase, the G2 phase is also a stage when the internal and external environments are monitored to ensure that faithful replication of the DNA has occurred and that conditions are favorable for cytokinesis. In addition, as for the G1 phase there is a major check-point at the end of the G2 phase that controls the entry into M-phase.
M-Phase
During M-phase there is an ordered series of events that leads to the alignment and separation of the duplicated chromosomes (called sister chromatids) This process is divided into distinct steps that were originally identified and characterized through light microscopic observations of dividing cells. The steps of mitosis are termed prophase, prometaphase, metaphase, anaphase and telophase. Although cytokinesis is the process by which the parental cell is physically separated into two new daughter cells, it actually begins during anaphase. The processes that occur during M-phase require much less time than those of S-phase, generally lasting only 1–2hrs.
- During prophase the duplicated chromosomes condense while outside the nucleus the mitotic spindle assembles between the two centrosomes. The centrosome is an organelle that serves as the main microtubule organizing center that is involved in the attachment of microtubules to the sister chromatids.
- During prometaphase the nuclear membrane breaks apart and the chromosomes can attach to spindle microtubules and begin active movement.
- During metaphase the chromosomes are aligned at the equator of the spindle midway between the spindle poles. The sister chromatids are attached to opposite poles of the spindle.
- During anaphase the sister chromatids synchronously separate to form the two sets of daughter chromosomes. Each sister chromatid is slowly pulled towards the spindle pole it faces.
- During telophase the two daughter chromosomes arrive at the spindle poles and decondense. A new nuclear envelope forms around each set of chromosomes which forms the two new nuclei. This process marks the end of mitosis and sets the stage for cytokinesis.
The Mechanics of Mitotic Cell Division
The process of mitotic cell division must occur in a highly ordered and accurate manner so as to ensure that each daughter cell receives an identical copy of the parental cells genome. Obviously the first step in this process is the accurate replication of the genome as described in the DNA: Chromatin Structure, Replication, DNA Damage Repair page. The processes described above relate to the regulation of the progressive steps taken as a cell progresses to cytokinesis. This section will discuss the biochemical processes undertaken to effect accurate separation of the duplicated DNA (the sister chromatids) and cytokinesis.
Following duplication of the chromosomes the sister chromatids are held together through multisubunit protein complexes referred to as cohesin. The cohesin complexes function in both mitotic and meiotic cell division. The human cohesin complex is composed of proteins encoded by eight different genes. These genes are STAG1 (stromal antigen 1), STAG2, STAG3, structural maintenance of chromosome 1A (SMC1A), SMC1B, SMC3, REC8 meiotic recombination protein (REC8), and RAD21 cohesin complex component (RAD21). The cohesin complexes are found all along the length of each chromatid as the DNA is replicated.
Following DNA replication the chromosomes condense and this is the role of proteins called condensins. The condensins are divided into two subgroups termed condensin I subunits and condensin II subunits. There are a total of eight human genes that encode condensin subunit I and condensin subunit II proteins. Two members of the structural maintenance of chromosome (SMC2 and SMC4) protein family are components of the condensins. The other six proteins in these complexes are referred to as non-SMC condensin complex (NCAP) subunits, three of which are condensin I subunits (NCAPD2, NCAPG, and NCAPH) and three of which are condensin II subunits (NCAPD3, NCAPG2, and NCAPH2). Chromosome condensation is the first easily identifiable sign that a cell is about to enter M-phase. Cohesins and condensins are structurally related and act in concert to prepare the chromosomes for mitosis.
The task of separating the sister chromatids, such that each daughter cell receives one copy of each chromosome, is carried out by the mitotic spindle which is composed of microtubules and numerous additional proteins that interact with them such as the kinesin and dynein molecular motors. Many of the additional proteins, that are collectively referred to as microtubule-associated proteins (MAP), are required for correct formation and orientation of the mitotic spindles.
An additional cytoskeletal structure is required for the actual separation of the cell into two new cells. This structure is referred to as the contractile ring. The contractile ring is so-called because under microscopy it looks like a ring-shaped structure just beneath the plasma membrane at the future site where a cell will separate during cytokinesis. The contractile ring is composed of actin and myosin, as well as several additional proteins. Many of these additional proteins are septins (a family of 13 GTP-binding proteins), formins (a family of 15 RHO-GTPase effector proteins), Arp2/3 complex [a complex of seven proteins including actin-related proteins 2 and 3 (ARP2 and ARP3)], tropomyosin, coronin (an actin-binding protein), anillin (an F-actin-binding protein), profilin (an actin-binding protein), a member of the IQ motif containing GTPase activating proteins (IQGAP), a myosin light chain kinase (MYLK), and Rho associated coiled-coil domain containing protein kinase 1 (ROCK1). The contractile ring assembles during anaphase and, as its name implies, it contracts as cells divide. In most, but not all, eukaryotic cell types, the contractile ring is responsible for cytokinesis.
The process of mitosis occurs through a highly ordered series of five steps as outlined above. The actual separation of the parental cell into two daughter cells can be considered the sixth step in mitosis. Whereas, prophase, prometaphase, metaphase, anaphase, and telophase occur in a strictly controlled sequential fashion, cytokinesis begins in anaphase and continues until the cell divides.
During interphase the microtubule machinery is in a constant state of dynamic instability. Individual microtubules are growing or shrinking at any given moment. During prophase the activation of M-phase cyclin/cyclin-dependent kinase (M-CDK) complexes initiates a change in the microtubule structures to one where there are a large number of shorter microtubules surrounding each centrosome. The centrosomes are cytoplasmic nucleation sites for the mitotic spindles. M-CDK complexes initiate these changes via the phosphorylation of microtubule motor proteins and microtubule-associated proteins (MAP).
During prometaphase the nuclear envelope abruptly breaks down as a consequence of M-CDK complexes phosphorylating lamin proteins of the nuclear lamina. The dissolution of the nuclear membrane allows the microtubules to access the mitotic spindles. When microtubules attach to the mitotic spindle they become stabilized. The microtubules eventually become attached at the kinetochore which is a complex protein structure that assembles onto the highly condensed DNA at the centromere. The chromosomes are pulled back and forth by the microtubules eventually becoming aligned equidistant from the two spindle poles. The alignment of the chromosomes forms the metaphase plate. The chromosomes oscillate about the metaphase plate awaiting the signal that will induce the sister chromatids to separate. This phase of mitosis is referred to as the spindle-attachment checkpoint and it ensures that cells do not enter anaphase until all of the chromosomes are attached to both poles of the mitotic spindles. As described above, sister chromatids begin to separate with the activation of the APC/C. As each sister chromatid is pulled towards one of the poles of the mitotic spindle the kinetochore microtubules depolymerize.
By the end of anaphase, the daughter chromosomes have separated to opposite ends of the cells and have begun to decondense which signals the onset of telophase. Telophase is denoted by the reassembly of the nuclear envelope around each group of daughter chromosomes. During this process the lamins that make up the nuclear lamina are dephosphorylated allowing them to re-associate with the nuclear envelope. Following the formation of the new nuclear envelopes, the chromosomes decondense into their interphase state and transcriptional activity begins anew. The cell is now ready for the final process, complete separation into two daughter cells.
Checkpoints and Cell Cycle Regulation
It should seem obvious that the processes that drive a cell through the cell cycle must be highly regulated so as to ensure that the resultant daughter cells are viable and each contains the complement of DNA found in the original parental cell. There are many “parts” to the systems that control the transit through a eukaryotic cell cycle. These “parts” include mechanisms to control the timing of events so that each individual process is turned on and off at the appropriate time, mechanisms to initiate each event in the correct order and to also ensure that each event is triggered only once per cell cycle, controls to ensure events occur in a linear, irreversible direction, redundancy, or back-ups to ensure the cycle functions properly even in the context of some malfunctioning parts, and systems that are adaptable so that cell cycle events can be modified in the context of different cell types and/or environmental conditions.
Many of the most important discoveries about the mechanisms that control events of the cell cycle were elucidated using yeasts which are single cell eukaryotes. By analysis of various mutants that inactivated genes encoding essential components of cell cycle control systems in yeast many important control genes were identified. These genes were identified as cell division cycle genes or cdc genes. Thus, many cell cycle control genes in mammalian cells are also called cdc genes. Much of the control of the progression through the phases of a cell cycle are exerted at checkpoints. There are many such checkpoints but the two most critical are those that occur near the end of G1 prior to S-phase entry and those near the end of G2 prior to mitosis.
Cyclins and Cyclin-Dependent Kinases
As indicated above, there is the need for cell cycle control mechanisms to exert their influences at specific times during each transit through a cell cycle. The heart of this timing control is the responsibility of a family of protein kinases that are called cyclin-dependent kinases, CDKs. The human genome contains a total of 26 genes that encode proteins of the CDK family. The kinase activity of these enzymes rises and falls as the cell progresses through a cell cycle. Different CDKs operate at different points in the cell cycle. As would be expected, the oscillating changes in the activity of CDKs leads to oscillating changes in the phosphorylation of various intracellular proteins. These phosphorylations alter the activity of the modified proteins which then effect changes in events of the cell cycle.
The cyclical activity of each CDK is controlled by a complex series of proteins, the most important of which are the cyclins, hence the name of the enzymes as cyclin-dependent kinases. The human genome contains a total of 30 genes that encode proteins of the cyclin family. The CDKs are absolutely dependent upon their interaction with the cyclins for activity. Unless they are tightly bound CDKs have no kinase activity. The cyclins were originally identified because they undergo a cycle of synthesis and degradation at specific points in each cell cycle. Thus, whereas the levels of the various CDKs remain fairly constant throughout the cell cycle, their activities change in concert with the fluctuations of the cyclins.
Four different classes of cyclins have been defined on the basis of the stage of the cell cycle in which they bind and activate CDKs. These four classes are G1-cyclins, G1/S-cyclins, S-cyclins, and M-cyclins. The following Table depicts the primary cyclins and CDK partners within these four mammalian classes.
Table of the Major Cyclin-CDK Complexes
| Cyclin-CDK Complex | Cyclin* | CDK Partner |
| G1-CDK | cyclin D | CDK4, CDK6 |
| G1/S-CDK | cyclin E | CDK2 |
| S-CDK | cyclin A | CDK2 |
| M-CDK | cyclin B | CDK1** |
*Humans express two cyclin A genes (CCNA1 and CCNA2), three cyclin B genes (CCNB1, CCNB2, and CCNB3), three cyclin D genes (CCND1, CCND2, and CCND3), and two cyclin E genes (CCNE1 and CCNE2). **CDK1 is the same as CDC2 in fission yeast and CDC28 in budding yeast.
The G1-cyclins are not found in all eukaryotic cells but in those where they are synthesized they promote passage through a restriction point in late G1 called Start. The G1/S-cyclins bind to their cognate CDKs at the end of G1 and it is this interaction that is required to commit the cell to the process of DNA replication in S-phase. The S-cyclins bind to their cognate CDKs during S-phase and it is this interaction that is required for the initiation of DNA synthesis. The M-cyclins bind to their cognate CDKs and in so doing promote the events leading up to and through cytokinesis.
CDK-Activating Kinases
Although CDKs are inactive unless bound to a cyclin, there is more to the activation process than just the interaction of the two parts of the complex. When cyclins bind to CDKs they alter the conformation of the CDK resulting in exposure of a domain that is the site of phosphorylation by a complex called CDK-activating kinase (CAK). Following CAK-mediated phosphorylation the cyclin-CDK complex is fully active. Humans express three genes encoding proteins in the CAK complex. These encoded proteins in the CAK complex are identified as cyclin H, cyclin dependent kinase 7, and MNAT1 component of CDK activating kinase. In addition to its role in the activation of several CDK-cyclin complexes the CAK complex associates with the TFIIH complex in the activation of RNA polymerase II.
CDK Inhibitors
In addition to control of CDK kinase activity by cyclin binding and CAK phosphorylation, control is exerted to inhibit CDK activity through interaction with inhibitory proteins as well as by inhibitory phosphorylation events. Thus, there is extremely tight control on the overall activity of each CDK. One of the inhibitory kinases that phosphorylates CDKs is called Wee1. The inhibitory phosphorylations are removed through the action of a phosphatase called CDC25. The action of these two regulatory enzymes on CDK activity is most important at the level of the M-CDK activity at the onset of the M-phase of the mitotic cell cycle. Proteins that bind to and inhibit cyclin-CDK complexes are members of the cyclin-dependent kinase inhibitor (CDKN) family. Mammalian cells express two subclasses of CDKN proteins. These are commonly referred to as CIP for CDK inhibitory proteins and INK4 for inhibitors of kinase 4. The CIP bind and inhibit CDK1, CDK2, CDK4, and CDK6 complexes, whereas the INK4 bind and inhibit only the CDK4 and CDK6 complexes.
There are at least three CIP proteins in mammalian cells and these are identified as p21Cip1/WAF1 (encoded by the CDKN1A gene), p27KIP1 (encoded by the CDKN1B gene), and p57KIP2 (encoded by the CDKN1C gene). The expression of each of these CIP genes is controlled by specific events that may have occurred during cell cycle transit. For example CDKN1A expression is induced in response to DNA damage. This induction is under the control of the action of the tumor suppressor protein p53 (see below).
There are at least four INK4 proteins (that are each identified by their molecular weights: p15INK4B (encoded by the CDKN2B gene), p16INK4A (encoded by the CDKN2A gene), and p18INK4C (encoded by the CDKN2C gene), and p19INK4D (encoded by the CDKN4D gene). The p15INK4B, p16INK4A, and p18INK4C proteins were the original three proteins of the INK class identified in humans. The p16INK4A protein is an important tumor suppressor since loss of its function is associated with a wide array of different cancer types. All the INK4 proteins contain four tandem repeats of a sequence of amino acids that were first identified in ankyrin and are thus referred to as ankyrin repeats.
Early Response Activation
As indicated above, many cells reside in a resting or quiescent state but can be stimulated by external signals to re-enter the cell cycle. These external growth promoting signals are the result of growth factors binding to their receptors. Most growth factors induce the expression of genes that are referred to as early and delayed-response genes. The activation of early response genes occurs in response to growth factor receptor-mediated signal transduction resulting in phosphorylation and activation of transcription factor proteins that are already present in the cell. Many of the induced early response genes are themselves transcription factors that in turn activate the expression of delayed-response genes. In the context of the cell cycle, these delayed-response genes encode proteins of the G1-CDK complexes.
One such early response gene is the proto-oncogene MYC. With respect to the cell cycle some of the genes turned on by activation of MYC are cyclin D, proteins of the ubiquitin ligase complex called SCF (Skp1/cullin/F-box protein) and the members of the E2F transcription factor family. There are six members of the E2F family: E2F1 through E2F6). The synthesis of cyclin D will result in the activation of G1-CDK complexes. The synthesis of components of SCF leads to the degradation of p27KIP1 which normally inhibits G1-CDK complexes. The synthesis of E2F family members results in increased synthesis of proteins involved in DNA synthesis as well as the synthesis of the S-phase cyclins A and E and CDK2. Regulation of E2F activity by the tumor suppressor pRB will be discussed below.
Anaphase Promoting Complex/Cyclosome, APC/C
The cyclical degradation of the cyclins is effected through the action of several different ubiquitin ligase complexes. The action of ubiquitin ligases in protein turn-over is discussed in more detail in the Protein, Organelle, and Cell Turnover page. There are two important ubiquitin ligase complexes that control the turn-over of cyclins and other cell cycle regulating proteins. One is the SCF complex which functions to control the transit from G1 to S-phase and the other is called anaphase promoting complex/cyclosome (APC/C) which controls the levels of the M-phase cyclins as well as other regulators of mitosis. The APC/C functions as a ubiquitin E3 ligase and is composed of 11-13 protein subunits. The catalytic core of the APC/C consists of the APC2 and APC11 encoded proteins which are members of two of the three major classes of E3 ligases in humans. APC2 is a cullin family protein and APC11 is RING domain (Really Interesting New Gene) containing protein.
One important function of APC/C is to control the initiation of sister chromatid separation which begins at the metaphase-anaphase transition. The attachment of the sister chromatids to the opposite poles of the mitotic spindles occurs early during mitosis. The ability of the sister chromatids to be pulled apart is initially inhibited because they are bound together by a cohesin protein complex. As discussed above, the cohesin complex is composed of eight protein subunits, three of which are members of the SMC (structural maintenance of chromosomes) family of proteins. The cohesin complex is deposited along the chromosomes as they are duplicated during S-phase.
Anaphase can only begin with the disruption of the cohesin complex. The breakdown of the cohesin complex is brought about as a consequence of the activation of the ubiquitin ligase activity of the APC/C. APC/C targets a protein that is the human homology of the yeast protein called securin. The human securin protein is encoded by the pituitary tumor-transforming 1 (PTTG1) gene. The PTTG1 encoded protein functions to inhibit the protease called separase (also called separin). The human separase protein is encoded by the extra spindle pole bodies like 1, separase (ESPL1) gene. The action of separase is to degrade the proteins of the cohesin complex, thus allowing sister chromatid separation.
Tumor Suppressors and Cell Cycle Regulation
Tumor suppressors are so called because cancer ensues as a result of a loss of their normal function, i.e. these proteins suppress the ability of cancer to develop. It would seem obvious, therefore, that one import function of tumor suppressors would be control of the progression of a cell through a round of the cell cycle. If cells are able to synthesize damaged DNA before it is repaired or to divide when the DNA is damaged then the resulting daughter cells can pass on the resultant DNA damage to their progeny. The result can be catastrophic resulting in cancer. For this reason, the two most important check points in the eukaryotic cell cycle are the G1-S transition and the entry into mitosis. The former prevents DNA replication prior to repair of damaged DNA and the latter prevents damage that may have occurred to the DNA during replication to propagated into daughter cells during mitosis.
Following the isolation and characterization of two tumor suppressor genes in particular it was found that they function to control the ability of cells to progress through these two important checkpoints. The protein encoded by the retinoblastoma susceptibility gene (pRB) and the p53 protein are both tumor suppressors. The function of pRB is to act as a brake preventing cells from exiting G1 and that of p53 is to inhibit progression from all phases of the cell cycle.
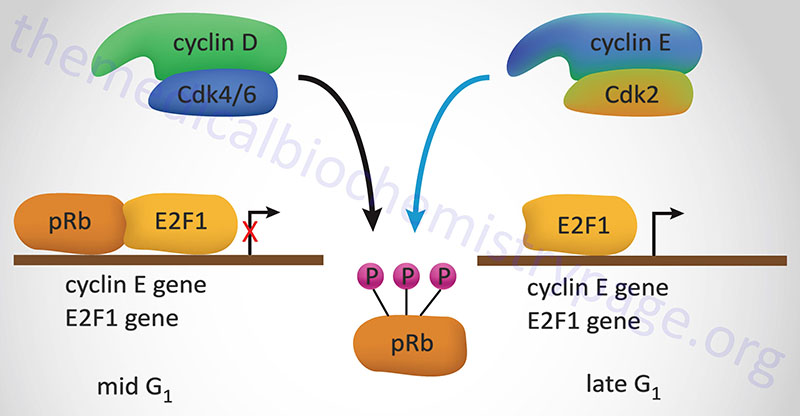
The best understood effect of G1-CDK activity is that exerted on transcription factors of the E2F family, hereafter referred to simply as E2F. In the context of the cell cycle regulation, E2F activates the expression of cyclin A, cyclin E and CDK2. These proteins are components of the S-CDK complexes necessary for progression through S-phase. The activity of E2F is itself controlled via interaction with pRB. When pRB binds E2F it can no longer function as a transcription factor as it is sequestered in the cytosol. Interaction of pRB and E2Fcorrelates to the state of phosphorylation of pRB and the affinity between the two proteins is highest when pRB is hypophosphorylated. Phosphorylation of pRB is maximal at the start of S phase and lowest after mitosis and entry into G1.
Stimulation of quiescent cells with mitogen induces phosphorylation of pRB, while in contrast, differentiation induces hypophosphorylation of pRB. One of the most significant substrates for phosphorylation by the G1 cyclin-CDK complexes is pRB. When pRB is phosphorylated by G1 cyclin-CDK complexes it releases E2F allowing E2F to transcriptionally activate its target genes. When E2F activates the expression of S-CDK complex proteins these complexes also target pRB for phosphorylation, thus maintaining the cell in a pro cell cycle progression state.
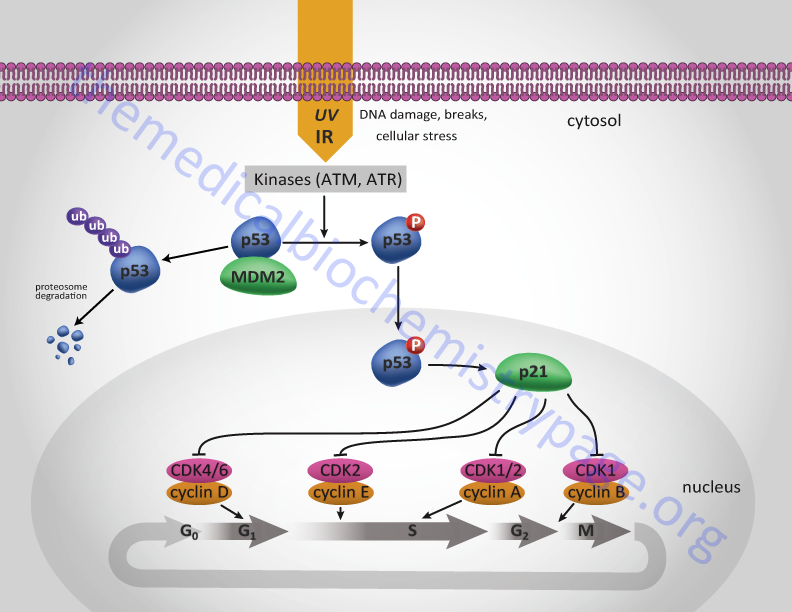
One major function of the p53 protein, which is active as a homotetrameric transcription factor, is to serve as a component of the checkpoint that controls whether cells enter as well as progress through S-phase. The action of p53 is induced in response to DNA damage. Under normal circumstances p53 levels remain very low due to its interaction with a member of the ubiquitin ligase family called MDM2. MDM2 is so named since it was isolated as an amplified gene in the tumorigenic mouse cell line 3T3DM. In response to DNA damage, e.g. as a result of uv-irradiation or γ-irradiation, cells activate several kinases including checkpoint kinase 2 (CHK2) and ataxia telangiectasia mutated (ATM).
One target of these kinases is p53. ATM also phosphorylates MDM2. When p53 is phosphorylated it is released from MDM2 and can carry out its transcriptional activation functions. One target of p53 is the cyclin inhibitor p21Cip1 gene. Activation of p21Cip1 leads to increased inhibition of the cyclin D1-CDK4 and cyclin E-CDK2 complexes thereby halting progression through the cell cycle either prior to S-phase entry or during S-phase. As a consequence of p53-induced synthesis of p21 expression, there is a convergence between the roles of p53 and pRB (as outlined above) in regulation of cyclin-CDK complexes. In either case the aim is to allow the cell to repair its damaged DNA prior to replication or mitosis.
Even given the limited discussion of the functions of pRB and p53 it is still easy to understand how loss of either protein function can lead to aberrant cell cycle progression and the potential for the development of cancer.
Introduction to Meiotic Cell Cycles
Humans, like most eukaryotes, reproduce sexually. Genetically this involves the mixing of the chromosomes from the haploid female gamete (ovum, egg) with those of the haploid male gamete (sperm). These specialized haploid cells are generated via the unique cell division process termed meiosis. The various stages of meiosis, both during meiosis I and meiosis II, are similar to those occurring during mitosis: prophase, metaphase, anaphase, and telophase.
The process of meiosis reduces the total chromosome number in the final cells (the gametes) by half. Meiosis reduces the chromosome number by half using many of the same molecular machines and control systems that operate in mitosis. The details of the steps of meiotic cell division will be addressed in the following sections but briefly, meiosis represents a process that is divided into two distinct events. In the first event, referred to as meiosis I, the DNA in the progenitor cell is duplicated and then the cell divides to produce two identical diploid cells. Since the duplicated chromosomes are separated in the final stages of meiosis I this stage is referred to as reductional cell division.
In the second event, referred to as meiosis II, the two diploid cells divide without first undergoing DNA replication such that the final results are the generation of haploid gametes (four sperm and one egg) from the originating diploid germ cell progenitor. Since the sister chromatids are separated in meiosis II, without prior DNA replication, this is referred to as equational cell division. Each diploid nucleus that enters meiosis produces four haploid nuclei, each of which contains either the maternal or paternal copy of each chromosome, but not both. The gamete progenitor cells are called oogonia in females and spermatogonia in males.
During male spermatogenesis the spermatogonia yield four functional haploid sperm. When oogonia undergo meiosis one of the two cells at the end of meiosis I represents the diploid cell that enters meiosis II and one of the resultant cells is referred to as a polar body, a small cell containing very little cytoplasm. The polar body then divides to become two polar bodies. During meiosis II the diploid cell (referred to as a secondary oocyte) divides, without DNA replication, to generate a functional gamete and another polar body. Thus, at the end result of female gametogenesis there are three polar bodies and one functional mature haploid ovum (egg). Polar bodies are not viable and do not mature into functional eggs.
The timing of the stages of meiosis in female gametes is dramatically different than in male gametes. Female eggs are arrested at the diplotene stage of prophase in meiosis I (see next section) until puberty when ovulation is stimulated. This means that female eggs have the potential to be in this arrested stage of meiosis for many years. Only eggs that undergo ovulation complete meiosis I prior to their release from the ovary. When the egg is fertilized only then does it complete meiosis II.
The Ordered Steps of Meiosis I
The DNA of meiotic cells is replicated during interphase to produce chromosomes that are composed of two sister chromatids. Even though there is twice as much DNA content in the meiotic cells at the end of DNA replication (referred to as 4C) the sister chromatids are still considered a single chromosome so that the chromosome number (referred to as 2N) is not changed at the end of meiotic DNA replication and the cell is, therefore, still technically a diploid cell.
During prophase of meiosis I (prophase I) the chromosomes condense, the nuclear membrane is degraded, and the homologous chromosomes form structures that are referred to as bivalents. The term bivalent chromosome refers to the tetrad structure that is formed as a result of the interaction of homologous regions of DNA between sister chromatids. The regions of homology that interact are the result of a multiprotein complex termed the synaptonemal complex (SC). In humans the SC is composed of core proteins encoded by the SYCP1, SYCP2, and SYCP3 genes and at least three additional proteins referred to as synaptonemal central element proteins.
The interaction of homologous DNA regions results in DNA double-strand breaks that are subsequently repaired via the process referred to as homology-directed repair. The exchange of DNA segments between the sister chromatids, one paternal and one maternal, occurs at sites in the chromosomes that are termed chiasma. This homologous recombination process results in gametes possessing chromosome that have both maternal and paternal sequences and represents a significant contribution to the evolution of humans.
The various substages of prophase I, in order, are leptotene (sister chromatids form visible strands in the nucleus), zygotene (the chromosome line up into homologous pairs), pachytene (fully formed tetrads), diplotene (homologous chromosomes separate), diakinesis (chromosome condensation occurs), and synchronous processes (centrosomes migrate to the two spindle poles).
During leptotene the homologous chromosomes condense and pair and the process of homologous recombination begins.
During zygotene, the synaptonemal complex (SC) assembles at sites where homologous recombination events are occurring.
The SC complex assembly is completed during pachytene and the homologous chromosomes are paired (synapsed) along their entire lengths.
Diplotene marks the stage of prophase I where the SC is disassembled and the chromosomes begin to condense with condensation completing during diakinesis. These latter stages of meiosis are the only times when it is possible to physically visualize cross-over events (chiasma) in the chromosomes. In female gametogenesis, meiosis halts at the diplotene stage of prophase I. This extended diplotene stage is referred to as dictyotene.
Following the completion of prophase I the cell enters metaphase I. During metaphase I the homologous pairs of chromosomes (the bivalents) move together along what is termed the metaphase plate in the middle of the cell. This movement is the result of microtubules of the kinetochores (the protein complex associated with the centromeres of the chromosomes) that are connected to the centrosomes (the nuclear organelle involved in formation of the spindle fibers).
During anaphase I the kinetochore microtubules contract resulting in the pulling of the homologously paired chromosomes to opposite poles. The cohesin complex that holds chromosome arms together is degraded but the cohesin complex at the centromere remains intact which results in the sister chromatids acting as a single chromosome and remaining together, only the homologs are separated.
During telophase I the spindle network is degraded and a new nuclear membrane may form (not always necessary) around each set of haploid chromosomes. Although these chromosomes are termed haploid, they contain the paired sister chromatids that joined during prophase I. The cell then undergoes cytokinesis generating two haploid daughter cells that are ready to undergo the stages of meiosis II.
The Ordered Steps of Meiosis II
As indicated above, meiosis II is the second meiotic division, and is designed to result in the separation of sister chromatids, a process referred to as equational segregation. The processes that are undertaken in meiosis II are very similar to those occurring in mitotic cells. Like mitosis, meiosis II occurs in four ordered steps identified, in order, as prophase II, metaphase II, anaphase II, and telophase II. Although similar, the genetic results of meiosis II and mitosis are fundamentally different.
The end result of meiosis II is production of four haploid cells from the two haploid cells that were derived through the process of meiosis I. It is important to understand that, in human gametes, at the end meiosis I, the two haploid cells have a chromosome number of 1 (designated 1N) where each chromosome consists of two sister chromatids. At the end of meiosis II the chromosome number is still 1 (1N) in the four haploid cells since the sister chromatids have been separated.
Prophase II begins as soon as the two daughter cells are formed at the end of meiosis I. During prophase II the chromosomes begin to condense once again which results in the shortening and thickening of the sister chromatids. The nuclear membrane dissolves and the centrosomes move to the polar regions to begin assembly of the spindle fibers that is required for the second meiotic division.
During metaphase II the chromosomes are connected to the centrioles through the microtubules of the kinetochores. The chromatids align resulting in the formation of the metaphase plate.
During anaphase II the centromeric cohesin which held the sister chromatids together during meiosis I is degraded. The loss of this cohesin allows the sister chromatids to segregate. At this stage in meiosis the sister chromatids are referred to as sister chromosomes and these chromosomes move toward opposing spindle poles.
Meiotic telophase II is similar to meiotic telophase I in that the chromosomes decondense and lengthen along with the disassembly of the spindle apparatus. The separation of the sister chromosomes results in the reformation of the nuclear membrane. Upon completion of the nuclear membrane formation the cells undergo cytokinesis resulting in the four haploid daughter cells. In males these four cells (sperm) are viable for fertilization. In females there are three polar bodies which are not functional gametes, and the one functional ovum (egg).
Meiotic Nondisjunction
The proper sorting of the chromosomes during meiosis must be highly precise such that at the end of the process one complete set of chromosomes is distributed to each of the four resulting haploid gametes. In order to properly sort the original 92 chromatids, that result from the duplication of each of the 46 chromosomes in the original diploid progenitor cells, there needs to be accurate separation, first of the sister chromatids and then of the sister chromosomes. This process of separation is termed disjunction.
Improper disjunction, termed nondisjunction, can occur either during meiosis I or during meiosis II. When nondisjunction occurs some of the resulting haploid gametes will lack a particular chromosome while others will end up with more than one copy. Nondisjunction is especially common in human female meiosis as a result of the many years the eggs remain arrested in meiosis I. Indeed, nondisjunction in meiosis I increases dramatically as female age advances. Chromosomal nondisjunction during egg development represents the most common cause of both spontaneous abortion and intellectual impairment in humans.
When nondisjunction occurs during meiosis I the result will be two disomic (containing two copies of a chromosome) gametes and two nullisomic (lacking a particular chromosome) gametes. Because monosomy for any autosome is non-viable the two nullisomic gametes, if used for fertilization, will not lead to any viable fetus. The gametes that contain the extra chromosome will lead to aneuploidy (any chromosome number that is not 46 or a multiple of an exact haploid number) and only a small set of aneuploid chromosomes result in viable embryos. Aside from trisomy of chromosomes 13, 18, or 21 no other autosomal aneuploidy is compatible with life. Trisomy 13 is the cause of Patau syndrome, trisomy 18 is the cause of Edward syndrome, and trisomy 21 is the cause of Down syndrome. Indeed, nondisjunction of chromosome 21 during female meiosis I represents the leading cause (75%–80%) of Down syndrome. Because all autosomal monosomy and the vast majority of autosomal aneuploidy are not compatible with life, the results of nondisjunction during meiosis I are almost always lethal.
When nondisjunction occurs during meiosis II the result will be two normal haploid gametes (euploid: meaning any exact multiple of 23 chromosomes), one disomic gamete, and one nullisomic gamete. Because there are two normal haploid gametes, 50% of the outcomes from nondisjunction during meiosis II are viable embryos. If nondisjunction of chromosome 21 occurs during meiosis II the one disomic gamete would lead to a Down syndrome offspring. Indeed, nondisjunction of chromosome 21 during meiosis II represents approximately 20% of Down syndrome patients.
