Last Updated: October 28, 2025
Introduction to the Ehlers-Danlos Syndromes
Ehlers-Danlos syndrome (EDS) represents a heterogeneous group of generalized connective tissue disorders. The predominant forms of EDS result from defects in collagen synthesis and or processing as well as several forms resulting from mutations in genes encoding enzymes in glycosaminoglycan processing or in the complement pathway of the immune system.
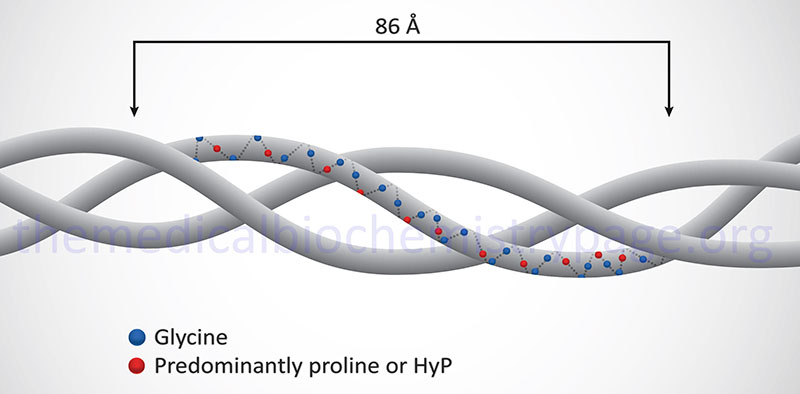
Collagen is the most abundant protein in the body and serves as a major building block of the extracellular matrix. Collagens are proteins that all contain three α-helical peptide chains wound in a triple helix. There are 44 expressed collagen genes dispersed throughout the human genome and the protein products from these genes combine to form 28 different types of triple helical collagen.
The major manifestations of the EDS family of disorders are skin fragility, skin hyperextensibility, and joint hypermobility. Original designations for the various forms of EDS used Roman numerals (e.g. type I, II, III, etc.). However, in 1997 the Villefranche nomenclature was adopted which uses descriptive designations, for example EDS type I now being referred to as classical EDS.
As of 2017 a total of 13 distinct forms of EDS had been characterized. Many of the EDS forms have significant overlapping clinical pathology. The predominant forms of EDS are classical EDS, vascular EDS, and hypermobile EDS. The other 10 recognized forms of EDS are considered rare and are classic-like EDS, kyphoscoliosis EDS, arthrochalasia EDS, dermatosparaxis EDS, periodontal EDS, brittle cornea syndrome EDS, musculocontractural EDS, spondylodysplastic EDS, myopathic EDS, and cardiac valvular EDS.
Clinical Features of Ehlers-Danlos Syndromes
Classic EDS
The originally identified EDS types I and II are now referred to as classical EDS. Classical EDS is the result of mutations in the collagen genes, COL5A1 and COL5A2. Classical EDS is inherited as an autosomal dominant disease. The characteristic clinical features of classical EDS are soft, velvety, and hyperextensible skin and easy bruising. The joints are quite hypermobile. Many patients with classical EDS have mitral valve prolapse. Trauma to the skin usually results in large gaping wounds that bleed less than expected. Repeated trauma to the knees, elbows, and shins leads to pigmented scarring. In addition, the skin develops thin “cigarette-paper” scars as a result of trauma.
Molecular Biology of Classic EDS
The COL5A1 gene is located on chromosome 9q34.3 and is composed of 67 exons that generate two alternatively spliced mRNAs encoding preproprotein isoform 1 (1838 amino acids) and preproprotein isoform 2 (1838 amino acids). Although both COL5A1 encoded isoforms are the same size, the isoform 2 preproprotein encoding mRNA contains an alternate exon in the 3′ coding region thus, the protein contains an alternate amino acid segment compared to the isoform 1 protein.
The COL5A2 gene is located on chromosome 2q32.2 and is composed of 61 exons that encode a 1499 amino acid preproprotein. Rarely, classical EDS is associated with COL1A1 mutations. Mutations in the COL1A1 gene are also associated with the four most common forms (types I-IV) of osteogenesis imperfecta.
Vascular EDS
Vascular EDS (formerly type IV) is the arterial type of the disease. Vascular EDS is inherited as an autosomal dominant disease and is the result of mutations in the COL3A1 genes. The mutations alter the synthesis, structure, and secretion of type III collagen. The characteristic features of vascular EDS are thin, translucent skin with visible veins which causes marked bruising. Patients with vascular EDS are subject to arterial, uterine, and bowel rupture.
Molecular Biology of Vascular EDS
The COL3A1 gene is located on chromosome 2q32.2 and is composed of 51 exons that generate two mRNAs from the use of alternative polyadenylation signals. The COL3A1 encoded preproprotein is 1466 amino acids in length. Rarely classical EDS is associated with COL1A1 mutations. Mutations in the COL1A1 gene are also associated with the four most common forms (types I-IV) of osteogenesis imperfecta.
Hypermobile EDS
Hypermobile EDS (formerly type III) is characterized by marked hypermobility of both the large and small joints. The skin is soft, smooth, velvety, and bruises easily but doesn’t scar like in classical EDS. A clear genetic mutation (or mutations) has not yet been characterized as being associated with the generation of hypermobile EDS. Within one family, characterized to have several members with hypermobile EDS, a mutation in the COL3A1 gene was identified. However, COL3A1 mutations are characteristic of vascular EDS and thus, it is likely that this family was incorrectly diagnosed.
Rare Forms of EDS
Classic-Like EDS
Classic-like EDS is an autosomal recessive form of EDS that exhibits features similar to that of classic EDS, hence the naming. Classic-like EDS is characterized by hyperextensible, velvety, easily bruised skin that does not lead to the atrophic scarring typical in classic EDS. In addition to skin pathology, classic-like EDS patients have generalized joint hypermobility with or without recurrent dislocations. Classic-like EDS results from mutations in the TNXB gene which encodes the tenascin-XB protein.
The TNXB gene is located on chromosome 6p21.33–p21.32 and is composed of 44 exons that generate three alternatively spliced mRNAs, each of which encode a distinct protein isoform.
Kyphoscoliosis EDS
Kyphoscoliosis EDS (formerly type VIA) is the ocular type of EDS. It is inherited as an autosomal recessive disease and results from deficiencies in lysine hydroxylation due to mutations in the procollagen-lysine 2-oxoglutarate 5-dioxygenase 1 (PLOD1) gene. The PLOD1 encoded enzyme is a member of the 2-oxoglutarate and Fe2+-dependent dioxygenase family (2OG-oxygenases) of enzymes. Patients with kyphoscoliosis EDS have ocular fragility in addition to soft, velvety, hyperextensible skin, and hypermobile joints.
The PLOD1 gene is located on chromosome 1p36.22 and is composed of 20 exons that generate two alternatively spliced mRNA encoding precursor proteins of 771 amino acids (isoform 1) and 727 amino acids (isoform 2).
Arthrochalasia EDS
Arthrochalasia EDS (formerly type VII) comprises two distinct forms (formerly identified as VIIA and VIIB) that are autosomal dominant disorders. These disorders get their designation from the prominent symptom of abnormal relaxation or flaccidity of the joints. Arthrochalasia EDS results from mutations in the COL1A1 gene (originally designated as the VIIA form) that eliminates the N-proteinase cleavage site. Mutations in the COL1A2 gene that eliminate the N-proteinase cleavage site are also defined as causing arthroscalasia EDS (formerly the VIIB type).
The COL1A1 gene is located on chromosome 17q21.33 and is composed of 51 exons that generate two mRNAs via the use of alternative polyadenylation signals, both of which encode the same 1464 amino acid preproprotein.
The COL1A2 gene is located on chromosome 7q21.3 and is composed of 52 exons that generate three mRNAs through the use of alternative polyadenylation signals. These mRNAs encode a preproprotein of 1366 amino acids.
Mutations in the COL1A1 and COL1A2 genes are also associated with the four most common forms (types I-IV) of osteogenesis imperfecta.
Dermatosparaxis EDS
Dermatosparaxis EDS (formerly type VIIC) is inherited as an autosomal recessive disorder that is characterized by loss of skin strength leading to easy tearing. Dermatosparaxis EDS results from deficiencies in procollagen I N-proteinase activity (NPI). The NPI activity hydrolyzes the N-propeptide from type I, II, and V procollagens. The NPI activity is a protein encoded by the ADAMTS2 gene (a disintegrin-like and metalloproteinase with thrombospondin type 1 motif, 2).
The ADAMTS2 gene is located on chromosome 5q35.3 and is composed of 25 exons that generate two alternatively spliced mRNAs that give rise to two isoforms of the enzyme.
Periodontal EDS
Periodontal EDS (formerly type VIII) is inherited as an autosomal dominant disease. Periodontal EDS can be distinguished from other forms of EDS by the manifestation of severe gingival recession and periodontitis which results in premature loss of permanent teeth and resorption of alveolar bone by the third decade of life. There is no vascular or organ rupture phenotype in periodontal EDS. Skin anomalies in periodontal EDS are similar to those found in classical EDS. Periodontal EDS results from mutations in the C1R and C1S genes.
The C1R gene encodes the proteolytic subunit of the complement system C1 complex. The C1R gene is located on chromosome 12p13.31 and is composed of 12 exons that generate two alternatively spliced mRNAs, both of which encode distinct protein isoforms.
The C1S gene encodes a serine protease that forms a complex with the C1R encoded protein and another component of the C1 complex encoded by the C1Q gene. The C1S gene is also located on chromosome 12p13.31 and is composed of 12 exons that generate three alternatively spliced mRNAs that collectively encode two distinct protein isoforms.
Brittle Cornea Syndrome
Brittle cornea syndrome (BCS) is an autosomal recessive disorder that is characterized by the progressively thinning of the cornea, early-onset progressive keratoglobus or keratoconus (protrusion and thinning of the cornea), nearsightedness, hearing loss, and blue sclerae. Symptoms characteristic of EDS such as hypermobile joints and hyperelastic skin, are also seen often in BCS patients. Brittle cornea syndrome is composed of two subtypes, BCS1 and BCS2. BCS1 was originally referred to as EDS type VIB.
BCS1 results from mutations in the ZNF469 gene which encodes the zinc finger protein 469. The ZNF469 gene is located on chromosome 16q24.2 and is composed of 5 exons that encode a 3953 amino acid protein.
BCS2 results from mutations in the PRDM5 gene which encodes the PR/SET domain-containing protein 5. The PRDM5 gene is located on chromosome 4q27 and is composed of 23 exons that generate five alternatively spliced mRNAs, each of which encodes a distinct protein isoform.
Musculocontractural EDS
Musculocontractural EDS is an autosomal recessive disorder characterized by congenital multiple contractures, characteristically adduction-flexion contractures and/or talipes equinovarus (clubfoot), characteristic craniofacial features, which are evident at birth or in early infancy, and skin features such as skin hyperextensibility, bruising, skin fragility with atrophic scars, and increased palmar wrinkling.
Musculocontractural EDS results from mutations in the CHST14 gene which encodes the dermatan-4-sulfotransferase-1 (D4ST1) enzyme. D4ST1 catalyzes the 4-O-sulfation of N-acetylgalactosamine (GalNAc) residues in dermatan sulfates.
The CHST14 gene is located on chromosome 15q15.1 and is an intronless gene that encodes a 376 amino acid protein.
Spondylodysplastic EDS
Spondylodysplastic EDS represents a family of three related autosomal recessive disorders.
EDS spondylodysplastic type 1 is caused by mutations in the B4GALT7 gene which encodes the β-1,4-galactosyltransferase 7 enzyme (also called galactosyltransferase 1). This B4GALT7 encoded enzyme is involved in the glycosaminoglycan-protein linkages in proteoglycans. EDS spondylodysplastic type 1 is characterized by short stature, muscle hypotonia (that can range from severe congenital to a later onset mild form), and bowing of the limbs. The B4GALT7 gene is located on chromosome 5q35.3 and is composed of 8 exons that encode a 327 amino acid protein.
EDS spondylodysplastic type 2 is caused by mutations in the B3GALT6 gene that encoded the β-1,3-galactosyltransferase 6 enzyme. The B3GALT6 encoded enzyme is involved in the synthesis of heparan sulfates and chondroitin sulfates. EDS spondylodysplastic type 2 is characterized by kyphoscoliosis, tapered fingers, osteoporosis, aortic aneurysm, and problems with the lungs. The B3GALT6 gene is located on chromosome 1p36.33 and is an intronless gene that encodes a 329 amino acid protein.
EDS spondylodysplastic type 3 results from mutations in the SLC39A13 gene. The SCL39A13 gene encodes a transporter of the solute carrier family, specifically a zinc transporter. EDS spondylodysplastic type 3 is characterized by patients with protuberant eyes, wrinkled palms of the hands, tapering fingers, and distal joint hypermobility. The SLC39A13 gene is located on chromosome 11p11.2 and is composed of 13 exons that generate three alternatively spliced mRNAs, each of which encode distinct protein isoforms.
Myopathic EDS
Myopathic EDS is characterized by congenital muscle hypotonia and/or muscle atrophy that improves with age, proximal joint contractures (joints of the knee, hip and elbow), and hypermobility of distal joints (joints of the ankles, wrists, feet and hands). Myopathic EDS results from mutations in the COL12A1 gene.
The COL12A1 gene is located on chromosome 6q13-q14.1 and is composed of 68 exons that generate two alternatively spliced mRNAs that encode a 3063 amino acid preproprotein (long isoform) and a 1899 amino acid preproprotein (short isoform).
Cardiac Valvular EDS
Cardiac valvular EDS is inherited as an autosomal recessive disease that is characterized by severe progressive cardiac-valvular problems (aortic valve, mitral valve), skin problems (hyperextensibility, atrophic scars, thin skin, easy bruising), and joint hypermobility (generalized or restricted to small joints). Cardiac valvular EDS results from mutations in the COL1A2 gene. Mutations in the COL1A2 gene are also associated with the four most common forms (types I-IV) of osteogenesis imperfecta.